Abstract
Purpose
Direct sequencing (DS) is the standard method for detection of epidermal growth factor receptor (EGFR) gene mutation in non-small cell lung cancer (NSCLC); however, low detection sensitivity is a problem. The aim of this study is to demonstrate higher detection rate of EGFR gene mutation with peptide nucleic acid (PNA) clamping compared with DS.
Materials and Methods
This is a single arm, prospective study for patients with stage IIIB/IV or relapsed NSCLC. Using tumor DNA from 138 patients, both DS and PNA clamping for EGFR gene in exon 18, 19, 20, and 21 were performed. Discrepant results between the two methods were verified using Cobas and a mutant enrichment based next generation sequencing (NGS). Patients with activating mutations were treated with EGFR tyrosine kinase inhibitor (EGFR-TKI, gefitinib, or erlotinib) as first line treatment.
Results
Of 138 paired test sets, 24 (17.4%) and 45 (32.6%) cases with activating mutations were detected by DS and PNA clamping, respectively. The difference of detection rate between the two methods was 15.2% (95% confidence interval, 8.7% to 17.8%; p < 0.001). Between the two methods, 25 cases showed discrepant results (n=23, PNA+/DS–; n=2, PNA–/DS+). Mutations were confirmed by Cobas or NGS in 22 of 23 PNA+/DS– cases. The response rates to EGFR-TKI were 72.2% in the PNA+/DS+ group and 85.0% in the PNA+/DS– group.
In randomized clinical trials, epidermal growth factor receptor–tyrosine kinase inhibitors (EGFR-TKIs) have been identified as the best choice of treatment over the conventional platinum doublet chemotherapy in advanced nonsmall cell lung cancer (NSCLC) with EGFR gene mutations [1-4]. In contrast, EGFR wild-type tumors respond better to conventional platinum doublet chemotherapy than to EGFR TKIs [1,2,5,6]. Thus, the EGFR mutant status in NSCLC is used to help guide treatment decisions, so that detection of EGFR mutantations is clinically important.
A number of methods for detection of EGFR gene mutations have been developed. Among them, direct sequencing (DS) is regarded as the standard method for detection of EGFR gene mutations. However, DS is complex and time-consuming when used as a routine screening method [7]. In addition, because DS can detect only mutant DNA that comprise at least 25% of the total DNA, it has low sensitivity compared to other methods [8].
In clinical settings, tissue or cytology samples often contain a small subpopulation of mutant cells mixed with a greater level of normal tissue, thus leading to non-detection of mutations by DS. Thus tumor cell enrichment with tissue microdissection is generally used for surgically acquired paraffin blocks. However, microdissection of tiny tissues acquired from bronchoscopy or needle biopsy and cytology specimens is not easily performed in daily clinical practice. Therefore, the necessity of more sensitive methods for detection of EGFR gene mutations has been emphasized.
Peptide nucleic acid (PNA) is an artificially synthesized DNA analog that binds strongly to its complementary DNA sequence. While the specifically designed PNA probe inhibits polymerase chain reaction (PCR) amplification in wild-type sequences, it allows greater amplification in mutant type sequences [9]. PNA clamping enables detection of EGFR gene mutations in samples containing as few as roughly 1% mutant alleles [8]. Other advantages of PNA clamping include simplicity and speed when applied in clinical settings, although it cannot detect novel mutations [10].
The aims of this study are to demonstrate higher detection rate of EGFR gene mutation with PNA clamping compared with DS and to verify the discrepant results for EGFR gene mutation between two methods using another real-time PCR technique and next generation sequencing (NGS).
This was a single arm, open label, prospective and observational study for detection of EGFR gene mutations by PNA clamping and DS methods. DS and PNA clamping for EGFR gene were performed using DNA acquired from tumor tissue or cytology specimens. This study was approved by the Institutional Review Board of the hospital (HCRI 12-084-3), and registered at clinicalTrials.gov (NCT01767974). Written informed consent was obtained from all patients enrolled in this trial.
This study included female or male patients, aged 18 years or over, with stage IIIB or IV or relapsed NSCLC. The patients had an Eastern Cooperative Oncology Group (ECOG) performance status of 0-2. Patients were excluded if they had a previous exposure to EGFR-TKIs or any evidence of severe uncontrolled systemic disease as judged by the investigator or other significant clinical disorders or laboratory findings undesirable for the subject to participate in this study. Patients without available tumor DNA and women with child-bearing potential were also excluded from this study.
From January 2013 to September 2013, 146 patients were recruited. Of 146 patients, three patients failed during the screening and five patients were dropped out because of insufficient samples (n=3) and non-lung cancer (n=2). Insufficient samples indicated that the amount of tissue was too small for analysis. Non-lung cancer included a case of uterine cervical cancer and another of cholangiocarcinoma. Finally, 138 patients were enrolled in this study, and formalin-fixed paraffin-embedded tissue biopsy or cytology specimens from these patients were collected. Using 138 paired samples, DS and PNA clamping for EGFR gene mutation were performed (Fig. 1).
Most of the enrolled patients were treated with EGFR-TKIs after detection of activating EGFR gene mutations by at least one method. Response to EGFR-TKIs was evaluated using ver. 1.1 of the Response Evaluation Criteria in Solid Tumors (RECIST) [11].
DNA was extracted from five paraffin sections (10 μm) of the tumor tissue or cell block cytology specimens. The paraffin sections acquired from surgical resections or excision biopsy of lymph nodes were microdissected to enrich tumor cells. However, tiny tissues acquired from bronchoscopic or needle biopsy and cytology specimens were not microdissected in order to reflect the daily clinical practice. Before DNA extraction, the tissue was deparaffinized in xylene and then washed in 70% ethanol. DNA was isolated using a Gene All Tissue DNA Purification Kit (General Biosystem, Seoul, Korea) according to the manufacturer’s protocol. The extracted DNA was stored at –20°C until use.
For PCR, three sets of previously reported primers [12] were used. PCRs were carried out with an initial 5-minute incubation at 94°C, followed by 40 cycles of 30 seconds at 94°C (denaturation), 30 seconds at 63°C (annealing), and 30 seconds at 72°C (extension), and then a final incubation at 72°C for 10 minutes. For one PCR reaction, 50 ng of template DNA was mixed with 1 μL of each primer at a concentration of 10 pmol, 0.75 units of Taq DNA polymerase (Solgent, Daejeon, Korea), and 0.2 mmol/L deoxynucleotide triphosphate. Exons 18, 19, 20, and 21 of the EGFR gene were sequentially amplified and the products were sequenced. The PCR products were purified using a PCR clean-up kit(Macherey-Nagel, Dueren, Germany). Sequencing was performed using a BigDye Terminator Kit (Applied Biosystems, Foster City, CA) and an ABI Prism 3100 DNA Analyzer (Applied Biosystems) according to the protocol of the manufacturer.
We used the PNA Clamp EGFR Mutation Detection Kit (Panagene Inc., Daejeon, Korea) for detection of EGFR gene mutations by real-time PCR. All reactions were performed in a volume of 20 μL with template DNA, a primer and PNA probe set, and fluorescence dye PCR master mix. Real-time PCR was performed using a CFX 96 Kit (Bio-Rad, Philadelphia, PA). PCR cycling conditions were carried out with a 5-minute hold at 94°C followed by 40 cycles of 94°C for 30 seconds, 70°C for 20 seconds, 63°C for 30 seconds, and then 72°C for 30 seconds. Detection of each of 29 EGFR gene mutations was possible via one-step PNA-mediated real-time PCR clamping [13]. The efficiency of PCR was determined by measuring the threshold cycle (Ct) values, which were automatically calculated from PCR amplification plots of fluorescence versus the number of cycles. The delta Ct (ΔCt-1) was calculated to ensure that the sample and standard Ct values were from the tested sample and wild-type DNA control sample. When the ΔCt-1 value (standard Ct value minus sample Ct value) was more than 2.0, the sample was considered as mutant status. When the ΔCt-1 value was between 0 and 2.0, the ΔCt-2 value (sample Ct value minus non-PNA Ct value) was calculated. The sample was considered as mutant status if ΔCt-2 value was less than 3.0.
The EGFR PCR test (Cobas EGFR Mutation Test, Roche Molecular Systems Inc., Branchburg, NJ) is a CE-IVD marked multiplex allele-specific PCR-based assay designed for detection of 41 mutations in exons 18, 19, 20, and 21 of the EGFR gene [14]. A minimum of 150 ng of genomic DNA is required for PCR amplification. The EGFR PCR test was performed using the Cobas z 480 Analyzer per manufacturer’s package insert, and results were analyzed automatically and reported. The limit of detection has been validated to 5% mutant alleles. The workflow from DNA isolation to results reporting can be performed in one 8-hour period.
In samples with wild-type or invalid results by Cobas, mutant enriched pyrosequencing (Insight Onco, SeaSun BioMaterials, Daejeon, Korea) was performed. We also performed an additional search for resistance mutations to EGFR-TKIs for cases that progressed to EGFR-TKI treatment despite activating mutation of EGFR gene. Those were T790M mutation in exon 20, Kirsten rat sarcoma viral oncogene homolog (KRAS) gene mutation, phosphoinositide-3-kinase catalytic alpha (PIK3CA) gene mutation and human epidermal growth factor receptor 2 (HER2) gene amplification.
Pyrosequencing was performed according to the manufacturer’s protocol for amplicons using the GS Junior System (Roche Diagnostics, Mannheim, Germany). Emulsion PCR, breaking, and bead enrichment were performed using the GS Junior Titanium emPCR Kit Lib-L, emPCR Reagents Lib-L kit, Oil and Breaking Kit, and the Bead Recovery Reagents Kit according to the supplier’s instructions (Roche Diagnostics). For emulsion PCR, we used a copy-per-bead ratio of 0.5. Enrichment of DNA-carrying beads was performed using magnetic beads and a magnetic particle collector (Invitrogen, Carlsbad, CA). To determine the number of enriched beads, the GS Junior Bead Counter (Roche Diagnostics) was used for counting. Finally, we loaded 500,000 beads onto the PicoTiterPlate (Roche Diagnostics).
Sequencing was performed according to standard Roche/454 protocols using the GS Titanium Sequencing Kit (Roche Diagnostics) and the GS Junior device. We aimed at obtaining approximately 1,000 to 2,000 sequence reads for conventional pyrosequencing and Insight Onco sequencing and 10,000 to 20,000 sequencing reads for ultra-deep sequencing from each sample. After ultra-deep pyrosequencing, the reads from different samples were identified by the sample-specific sequence tags.
We hypothesized that PNA clamping would be at least 10% more sensitive than DS. A sample size of 140 pairs was calculated considering 20% drop-out rate and 16% discordant pairs in order to have 90% power to detect a difference in proportions of 0.1. McNemar’s test with a 0.05 one-sided significance level was used for comparison of detection rate for EGFR gene mutation between PNA clamping and DS. Concordance between the two methods was determined using Cohen's kappa statistics. A two-sided p-value less than 0.05 indicated statistical significance except McNemar’s test. All analyses were performed using IBM SPSS ver. 20.0 (IBM Co., Armonk, NY).
The baseline characteristics of the 138 enrolled patients and specimens are summarized in Table 1. The mean age of the patients was 65.8 years. Most patients were male (64.5%), and adenocarcinoma (89.9%) was the major histologic type followed by squamous cell carcinoma (7.2%). Patients who had never smoked accounted for 46.4% of the study population.
Of all specimens analyzed, 112 (81.2%) were biopsy samples and 26 (18.8%) were cytology samples from cell blocks. Most biopsy samples were obtained from lung (54.3%) via bronchoscopic mucosal biopsy, bronchoscopic transbronchial lung biopsy, surgery, endobronchial ultrasound guided transbronchial needle aspiration, and computed tomography guided transthoracic needle biopsy. The rest of them were obtained from lymph node, bone, brain, and adrenal gland. Among cytology samples, 11 were pleural fluid and 15 were bronchial brushing.
Activating mutations of EGFR gene were detected in 45 (32.6%) and 24 (17.4%) of 138 samples by PNA clamping and DS, respectively (Table 2). In 22 cases, the same activating mutations were detected by both methods, while discrepant results between them were shown in 25 cases (PNA+/DS–, 23 cases; PNA–/DS+, 2 cases). Difference of detection rate for EGFR gene mutation was 15.2% (95% confidence interval, 8.7% to 17.8%; p < 0.001). Thus, PNA clamping was more sensitive than DS for detection of EGFR gene mutation.
Of the 45 mutations detected by PNA clamping, 26 (57.8%) were in-frame deletions in exon 19, 18 (40.0%) were point mutations (L858R or L861Q) in exon 21, and only one point mutation (G719X) was detected in exon 18. By contrast, EGFR gene mutations were detected in only 24 cases by DS, including 10 (41.7%) in-frame deletions in exon 19, 13 (54.2%) point mutations in exon 21, and one point mutation in exon 18 (Table 2).
Of the 138 cases, 113 cases showed overall agreement between PNA clamping and DS, consisting of 22 cases with activating mutations and 91 cases without activating mutations. Among the 91 cases without activating mutations, PNA clamping found one case with exon 20 insertion mutation and DS detected 27 cases with germ line single nucleotide polymorphisms (Table 2).
Among 25 discordant cases, two mutations detected by DS (exon 21 L858R) were reported as wild type by PNA clamping. Twenty-three mutations detected by PNA clamping (16 cases of exon 19 deletion and 7 cases of exon 21 L858R/L861Q) were not identified by DS. The concordance between both methods was 81.9% (κ coefficient=0.62, p < 0.001).
Of the 47 patients with EGFR gene mutations detected by PNA clamping or DS, 41 cases were treated with EGFR-TKIs. Except for one patient who stopped taking EGFR-TKIs due to severe adverse events, among the 40 response evaluable patients, 30 patients showed partial responses, seven patients showed stable disease (SD), and three patients showed progressive disease (PD). The overall response rate (ORR) was 75.0% (30/40) and the disease control rate was 92.5% (37/40) (Table 3).
We divided 40 patients with EGFR gene mutations, who received EGFR-TKIs treatment into three subgroups, including PNA+/DS+ (n=18), PNA+/DS– (n=20), and PNA–/DS+ (n=2). The ORR was 72.2% (13/18) in the PNA+/DS+ group, and 85.0% (17/20) in the PNA+/DS– group. There was one increasing SD, and one PD in two PNA–/DS+ patients (Table 3).
Validation results of the 25 cases (PNA+/DS–, n=23; PNA–/DS+, n=2) are summarized in Table 4. In cases with wild type or invalid results by Cobas, NGS was performed. When one of the two validation methods showed an activating mutation, validation result was defined as having an activating mutation, whereas, validation result was defined as wild type when both methods showed wild-type.
Among the 23 PNA+/DS– cases, validation methods found the same mutations in 22 cases, except one case with insufficient DNA. Two patients who showed PD with EGFRTKI proved to have either HER2 amplification or K-ras and PIK3CA mutations, which are implicated with EGFR-TKI resistance. Of the two PNA–/DS+ patients, validation found the same mutation in one case, but wild type in the other case. A resistance mutation, PIK3CA, was found in one case that showed PD with EGFR-TKI.
Predictive values of PNA clamping for detection of activating mutations of the EGFR gene are summarized in Table 5. Of the 93 PNA– cases, only one case had an activating mutation, thus the negative predictive value was 98.9% (92/93). Among the 45 PNA+ cases, 44 proved to have activating mutation with sequencing or other validation methods. Thus, the positive predictive value of PNA clamping was 97.8% (44/45).
The most common mutation associated with sensitivity to EGFR-TKIs in exon 18 to 21 is in-frame deletions in exon 19, followed by a point mutation (L858R) in exon 21. Other less common mutations include point mutation in exon 18 (G719X) and exon 21 (L861Q). Representative resistant mutations to EGFR-TKIs include a point mutation (T790M) and in-frame insertions in exon 20 [7]. In this study, exon 19 deletion and exon 21 point mutations accounted for approximately 96% of all EGFR gene mutations.
Although a variety of methods for detection of EGFR gene mutations has been developed and used in practice over recent years, there was no consensus on which method is most efficacious. Among them, despite its low sensitivity, DS is regarded as the current standard method for detection of EGFR gene mutations and discovery of novel mutations. A number of alternative methods for mutation detection with high sensitivity have been developed and used clinically.
In this study, we compared PNA clamping with DS for detection of EGFR gene mutations using 138 paired samples obtained from tissue biopsy and cytology. As expected, the detection rate of PNA clamping was significantly higher than that of DS. For cases with discrepant results between DS and PNA clamping, we also performed validation tests using Cobas or NGS to exclude the possibility of false positivity. Cobas is another real time PCR technique with high sensitivity and accuracy, which acquired CE-IVD and US-IVD. Like PNA clamping, Cobas cannot detect novel mutations other than pre-designed mutations of the EGFR gene, thus NGS was performed in order to find novel or known resistance mutations.
Among the 23 cases with PNA+/DS– results, 22 cases were verified to have the same activating mutations as those detected by PNA clamping. Thus, we demonstrated that PNA clamping is a highly sensitive method with high positive and negative predictive values for detection of EGFR gene mutations compared with DS using clinical samples obtained from patients with advanced NSCLC.
In a previous study [15] using mostly surgically acquired tissues, sensitivity of DS was similar to that of PNA clamping, thus emphasizing the importance of enrichment method (tissue microdissection). Significantly lower sensitivity of DS in the current study can be caused by the low proportion (< 25%) of tumor cells in PNA+/DS– cases. However, 10 of 23 PNA+/DS– cases were surgically resected specimens on which we performed tissue microdissection to enrich tumor DNA. This result suggests that even in enriched tumor DNA, EGFR mutation containing DNA can occupy a minor proportion (intratumor heterogeneity). Thus, in view of the difficulties in performance of tissue microdissection in every clinical specimen, and considering intratumor heterogeneity, more sensitive detection methods than DS are preferred.
Another study [9] comparing DS and PNA clamping using cell blocks of malignant pleural effusion also reported a higher detection rate of PNA clamping (39%, 16/41) compared with DS (14.6%, 6/41). In their study, PNA clamping tended to be more sensitive in cell blocks containing a low proportion of tumor cells; this finding also supports the merit of sensitive detection methods.
Because the DNA from cytology samples may have a higher proportion of tumor DNA compared to DNA from biopsy samples, we also looked for any difference in the sensitivity of DS and PNA clamping separately in biopsy or cytology samples (Table 5). Although the sensitivities of DS and PNA clamping were numerically higher in cytology compared to biopsy samples, the difference of sensitivities between the methods were greater than 15% in both biopsy and cytology samples.
The ORR to EGFR-TKIs in patients with NSCLC harboring activating EGFR gene mutations was 75.0%, similar to that of previous studies [1,3,4,16]. In view of intratumor heterogeneity, sensitive techniques may detect minor tumor cell populations with EGFR mutantations, resulting in low clinical response to EGFR-TKIs. However, this was not the case in this study. In 20 patients with PNA+/DS–, the ORR was 85.0%, which is numerically higher compared to 65% of ORR in DS+ cases.
Among the PNA+/DS– group, two cases that showed progressive disease to EGFR-TKI treatment have other mutations leading to resistance to EGFR-TKIs. Among the resistant mutations, the most common mutation is the point mutation (T790M) in exon 20 [17]. Other mechanisms of resistance involve activating mutations of the main EGFR pathway, i.e., KRAS and PIK3CA gene mutations and HER2 gene amplification [18,19]. The mechanisms of EGFR-TKI resistance and treatment strategies are described in a review article [20].
In conclusion, PNA clamping is a highly sensitive and accurate method for detection of EGFR gene mutations, and the activating mutations detected by PNA clamping were translated to clinical responses. Because the higher sensitivity of PNA clamping was not compromised by the loss of predictive power of response to EGFR-TKI, we recommend use of sensitive EGFR mutation detection techniques other than DS in clinical settings.
References
1. Fukuoka M, Wu YL, Thongprasert S, Sunpaweravong P, Leong SS, Sriuranpong V, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS). J Clin Oncol. 2011; 29:2866–74.

2. Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010; 11:121–8.

3. Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011; 12:735–42.

4. Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012; 13:239–46.
5. Tsao MS, Sakurada A, Cutz JC, Zhu CQ, Kamel-Reid S, Squire J, et al. Erlotinib in lung cancer: molecular and clinical predictors of outcome. N Engl J Med. 2005; 353:133–44.
6. Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010; 362:2380–8.

8. Pao W, Ladanyi M. Epidermal growth factor receptor mutation testing in lung cancer: searching for the ideal method. Clin Cancer Res. 2007; 13:4954–5.
9. Han HS, Lim SN, An JY, Lee KM, Choe KH, Lee KH, et al. Detection of EGFR mutation status in lung adenocarcinoma specimens with different proportions of tumor cells using two methods of differential sensitivity. J Thorac Oncol. 2012; 7:355–64.

10. Kim HJ, Kim WS, Shin KC, Lee GH, Kim MJ, Lee JE, et al. Comparative analysis of peptide nucleic acid (PNA)-mediated real-time PCR clamping and DNA direct sequencing for EGFR mutation detection. Tuberc Respir Dis. 2011; 70:21–7.

11. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009; 45:228–47.

12. Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004; 350:2129–39.

13. Kim HJ, Lee KY, Kim YC, Kim KS, Lee SY, Jang TW, et al. Detection and comparison of peptide nucleic acid-mediated real-time polymerase chain reaction clamping and direct gene sequencing for epidermal growth factor receptor mutations in patients with non-small cell lung cancer. Lung Cancer. 2012; 75:321–5.

14. O'Donnell P, Ferguson J, Shyu J, Current R, Rehage T, Tsai J, et al. Analytic performance studies and clinical reproducibility of a real-time PCR assay for the detection of epidermal growth factor receptor gene mutations in formalin-fixed paraffin-embedded tissue specimens of non-small cell lung cancer. BMC Cancer. 2013; 13:210.
15. Lee HJ, Xu X, Kim H, Jin Y, Sun P, Kim JE, et al. Comparison of direct sequencing, PNA clamping-real time polymerase chain reaction, and pyrosequencing methods for the detection of EGFR mutations in non-small cell lung carcinoma and the correlation with clinical responses to EGFR tyrosine kinase inhibitor treatment. Korean J Pathol. 2013; 47:52–60.
16. Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009; 361:947–57.

17. Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013; 19:2240–7.

18. Ludovini V, Bianconi F, Pistola L, Chiari R, Minotti V, Colella R, et al. Phosphoinositide-3-kinase catalytic alpha and KRAS mutations are important predictors of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in patients with advanced non-small cell lung cancer. J Thorac Oncol. 2011; 6:707–15.

Fig. 1.
CONSORT diagram. NSCLC, non-small cell lung cancer; DS, direct sequencing; PNA, peptide nucleic acid clamping; NGS, next generation sequencing; EGFR-TKI, epidermal growth factor receptor–tyrosine kinase inhibitor.
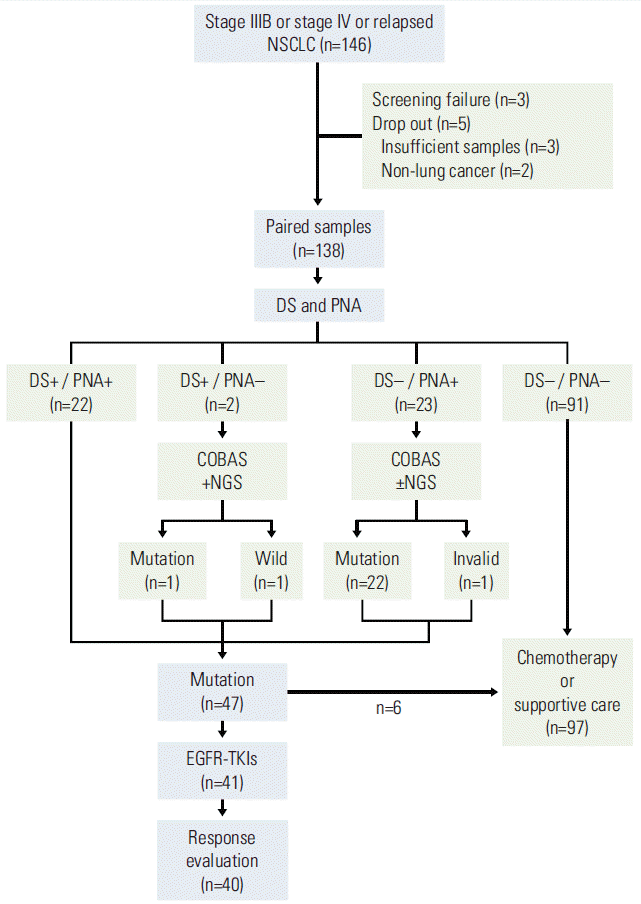
Table 1.
Characteristics of patients and specimens
| Variable | No. (%) |
|---|---|
| Age (yr) | |
| Mean±SD | 65.8±11.8 |
| Median (range) | 67.0 (23-87) |
| Gender | |
| Male | 89 (64.5) |
| Female | 49 (35.5) |
| Smoking status | |
| Never-smokers | 64 (46.4) |
| Former-smokersa) | 35 (25.4) |
| Current-smokers | 39 (28.2) |
| Histologic type | |
| Adenocarcinoma | 124 (89.9) |
| Squamous cell carcinoma | 10 (7.2) |
| Large cell carcinoma | 4 (2.9) |
| Specimen type | |
| Tissue biopsy | 112 (81.2) |
| Lung | 75 (54.3) |
| Bronchoscopic mucosal biopsy | 29 (21.0) |
| Bronchoscopic transbronchial lung biopsy | 11 (8.0) |
| Surgery | 28 (20.3) |
| EBUS-TBNA | 6 (4.3) |
| Computed tomography-guided needle biopsy | 1 (0.7) |
| Lymph node | 29 (21.0) |
| EBUS-TBNA | 21 (15.2) |
| Excisional biopsy | 8 (5.8) |
| Bone (rib, spine) | 3 (2.2) |
| Brain | 4 (3.0) |
| Right adrenal gland | 1 (0.7) |
| Cytology | 26 (18.8) |
| Pleural fluid | 11 (8.0) |
| Bronchial brushing | 15 (10.8) |
| Total | 138 (100) |
Table 2.
Agreement of PNA clamping and direct sequencing
| Variable |
EGFR gene mutation (PNA) |
Wild type or othera) | Total | Agreement | |||
|---|---|---|---|---|---|---|---|
| Exon 18 G719X | Exon 19 del | Exon 21 L858R/L861Q | |||||
| EGFR gene mutation (direct sequencing) | Exon 18 G719X | 1 | - | - | - | 1 | 0.819 (κ=0.62, p < 0.001) |
| Exon 19 del | - | 10 | - | - | 10 | ||
| Exon 21 L858R | - | - | 11 | 2 | 13 | ||
| Exon 21 L861Q | - | - | - | - | - | ||
| Wild type or SNPb) | - | 16 | 7 | 91a),b) | 114 | ||
| Total | 1 | 26 | 18 | 93 | 138 | ||
Table 3.
Response to EGFR–tyrosine kinase inhibitors in EGFR activating mutation positive cases
| Variable | PNA + | DS + | Anya) + | PNA +/DS + | PNA +/DS – | PNA –/DS + |
|---|---|---|---|---|---|---|
| Complete response | 0 | 0 | 0 | 0 | 0 | 0 |
| Partial response | 30 (78.9) | 13 (65.0) | 30 (75.0) | 13 (72.2) | 17 (85.0) | 0 |
| Stable disease | 6 (15.8) | 6 (30.0) | 7 (17.5) | 5 (27.8) | 1 (5.0) | 1 (50.0) |
| Progressive disease | 2 (5.3) | 1 (5.0) | 3 (7.5) | 0 | 2 (10.0) | 1 (50.0) |
| Total | 38 (100) | 20 (100) | 40 (100) | 18 (100) | 20 (100) | 2 (100) |
Table 4.
Validation of discrepant results between direct sequencing and PNA clamping
| No. | Age (yr) | Gender |
EGFR gene mutation |
Validation methods |
Resistance mechanismsa) | Response | ||
|---|---|---|---|---|---|---|---|---|
| Direct sequencing | PNA | Cobas | NGS | |||||
| 1 | 62 | M | Wild | L858R/L861Q | Wild | L858R | HER2 amplification | PD |
| 2 | 66 | M | Wild | L858R/L861Q | Wild | L858R | KRAS, PIK3CA mutation | PD |
| 3 | 57 | M | Wild | L858R/L861Q | Wild | L858R | - | PR |
| 4 | 66 | M | Wild | L858R/L861Q | L858R | - | - | PR |
| 5 | 53 | M | Wild | L858R/L861Q | L858R | - | - | NE |
| 6 | 63 | F | Wild | L858R/L861Q | L858R | - | - | SD |
| 7 | 56 | F | Wild | L858R/L861Q | Invalidb) | L858R | - | PR |
| 8 | 55 | M | Wild | E19 del | Invalidc) | Invalidc) | - | PR |
| 9 | 73 | F | Wild | E19 del | Invalidb) | E19 del | - | PR |
| 10 | 76 | F | Wild | E19 del | E1 | - | - | NE |
| 11 | 66 | F | Wild | E19 del | E1 | - | - | PR |
| 12 | 74 | F | Wild | E19 del | E1 | - | - | PR |
| 13 | 65 | M | Wild | E19 del | E1 | - | - | PR |
| 14 | 75 | M | Wild | E19 del | E1 | - | - | PR |
| 15 | 76 | F | Wild | E19 del | E1 | - | - | PR |
| 16 | 84 | F | Wild | E19 del | E1 | - | - | PR |
| 17 | 58 | F | Wild | E19 del | E1 | - | - | PR |
| 18 | 60 | F | Wild | E19 del | E1 | - | - | SD |
| 19 | 57 | F | Wild | E19 del | E1 | - | - | PR |
| 20 | 66 | M | Wild | E19 del | E1 | - | - | PR |
| 21 | 84 | M | Wild | E19 del | E1 | - | - | NE |
| 22 | 75 | F | Wild | E19 del | E1 | - | - | PR |
| 23 | 63 | M | Wild | E19 del | E1 | - | - | PR |
| 24 | 68 | M | L858R | Wild | Wild | L858R | PI3KCA | PD |
| 25 | 84 | M | L858R | Wild | Wild | Wild | None | SDd) |
Table 5.
Predictive values of PNA clamping for EGFR gene mutations
| DS |
PNA |
Sensitivity (%) |
|||
|---|---|---|---|---|---|
| Positive | Negative | DS | PNA | ||
| All samples (n=138) | Positive | 22 | 2 | 17.4 | 32.6 |
| Negative | 23 | 91 | |||
| Biopsy (n=112) | Positive | 18 | 1 | 17.0 | 32.1 |
| Negative | 18 | 75 | |||
| Cytology (n=26) | Positive | 4 | 1 | 19.2 | 34.6 |
| Negative | 5 | 16 | |||
| PPV and NPV of PNA | 97.8a) | 98.9b) | |||
| DS+Cobas or next generation sequencing | Positive | 44c) | 1d) | ||
| Negativee) | 1d),e) | 92c) | |||




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download