1. Goto Y, Yue L, Yokoi A, Nishimura R, Uehara T, Koizumi S, et al. A novel single-nucleotide polymorphism in the 3'-untranslated region of the human dihydrofolate reductase gene with enhanced expression. Clin Cancer Res. 2001; 7:1952–1956. PMID:
11448909.
2. Selga E, Noé V, Ciudad CJ. Transcriptional regulation of aldo-keto reductase 1C1 in HT29 human colon cancer cells resistant to methotrexate: role in the cell cycle and apoptosis. Biochem Pharmacol. 2008; 75:414–426. PMID:
17945194.

3. Serra M, Reverter-Branchat G, Maurici D, Benini S, Shen JN, Chano T, et al. Analysis of dihydrofolate reductase and reduced folate carrier gene status in relation to methotrexate resistance in osteosarcoma cells. Ann Oncol. 2004; 15:151–160. PMID:
14679136.

4. Padmanabhan S, Tripathi DN, Vikram A, Ramarao P, Jena GB. Methotrexate-induced cytotoxicity and genotoxicity in germ cells of mice: intervention of folic and folinic acid. Mutat Res. 2009; 673:43–52. PMID:
19110071.

5. Izbicka E, Diaz A, Streeper R, Wick M, Campos D, Steffen R, et al. Distinct mechanistic activity profile of pralatrexate in comparison to other antifolates in in vitro and in vivo models of human cancers. Cancer Chemother Pharmacol. 2009; 64:993–999. PMID:
19221750.

6. Affleck JG, Nowickyj SM, Walker VK. Selection for methotrexate resistance in mammalian cells bearing a Drosophila dihydrofolate reductase transgene: methotrexate resistance in transgenic mammalian cells. Cell Biol Toxicol. 2010; 26:117–126. PMID:
19337845.
7. Yang R, Li WW, Hoang BH, Kim H, Banerjee D, Kheradpour A, et al. Quantitative correlation between promoter methylation and messenger RNA levels of the reduced folate carrier. BMC Cancer. 2008; 8:124. PMID:
18452618.

8. Scionti I, Michelacci F, Pasello M, Hattinger CM, Alberghini M, Manara MC, et al. Clinical impact of the methotrexate resistance-associated genes C-MYC and dihydrofolate reductase (DHFR) in high-grade osteosarcoma. Ann Oncol. 2008; 19:1500–1508. PMID:
18385200.

9. Dulucq S, St-Onge G, Gagné V, Ansari M, Sinnett D, Labuda D, et al. DNA variants in the dihydrofolate reductase gene and outcome in childhood ALL. Blood. 2008; 111:3692–3700. PMID:
18096764.

10. Ongaro A, De Mattei M, Della Porta MG, Rigolin G, Ambrosio C, Di Raimondo F, et al. Gene polymorphisms in folate metabolizing enzymes in adult acute lymphoblastic leukemia: effects on methotrexate-related toxicity and survival. Haematologica. 2009; 94:1391–1398. PMID:
19648163.

11. Jansen G, Westerhof GR, Jarmuszewski MJ, Kathmann I, Rijksen G, Schornagel JH. Methotrexate transport in variant human CCRF-CEM leukemia cells with elevated levels of the reduced folate carrier. Selective effect on carrier-mediated transport of physiological concentrations of reduced folates. J Biol Chem. 1990; 265:18272–18277. PMID:
2211701.

12. Backus HH, Pinedo HM, Wouters D, Padrón JM, Molders N, van Der Wilt CL, et al. Folate depletion increases sensitivity of solid tumor cell lines to 5-fluorouracil and antifolates. Int J Cancer. 2000; 87:771–778. PMID:
10956384.

13. Marshall LA, Rhee MS, Hofmann L, Khodjakov A, Schneider E. Increased lysosomal uptake of methotrexate-polyglutamates in two methotrexate-resistant cell lines with distinct mechanisms of resistance. Biochem Pharmacol. 2005; 71:203–213. PMID:
16263093.

14. Hattinger CM, Stoico G, Michelacci F, Pasello M, Scionti I, Remondini D, et al. Mechanisms of gene amplification and evidence of coamplification in drug-resistant human osteosarcoma cell lines. Genes Chromosomes Cancer. 2009; 48:289–309. PMID:
19105235.

15. Gomes CM, van Paassen H, Romeo S, Welling MM, Feitsma RI, Abrunhosa AJ, et al. Multidrug resistance mediated by ABC transporters in osteosarcoma cell lines: mRNA analysis and functional radiotracer studies. Nucl Med Biol. 2006; 33:831–840. PMID:
17045162.

16. Kang MH, Harutyunyan N, Hall CP, Papa RA, Lock RB. Methotrexate and aminopterin exhibit similar in vitro and in vivo preclinical activity against acute lymphoblastic leukaemia and lymphoma. Br J Haematol. 2009; 145:389–393. PMID:
19298590.
17. Rothem L, Ifergan I, Kaufman Y, Priest DG, Jansen G, Assaraf YG. Resistance to multiple novel antifolates is mediated via defective drug transport resulting from clustered mutations in the reduced folate carrier gene in human leukaemia cell lines. Biochem J. 2002; 367:741–750. PMID:
12139489.

18. Patiño-García A, Zalacaín M, Marrodán L, San-Julián M, Sierrasesúmaga L. Methotrexate in pediatric osteosarcoma: response and toxicity in relation to genetic polymorphisms and dihydrofolate reductase and reduced folate carrier 1 expression. J Pediatr. 2009; 154:688–693. PMID:
19159907.

19. Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell. 2001; 7:627–637. PMID:
11463387.

20. Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000; 19:94–102. PMID:
10619848.

21. Weisman MH, Furst DE, Park GS, Kremer JM, Smith KM, Wallace DJ, et al. Risk genotypes in folate-dependent enzymes and their association with methotrexate-related side effects in rheumatoid arthritis. Arthritis Rheum. 2006; 54:607–612. PMID:
16447238.

22. Gellekink H, Blom HJ, van der Linden IJ, den Heijer M. Molecular genetic analysis of the human dihydrofolate reductase gene: relation with plasma total homocysteine, serum and red blood cell folate levels. Eur J Hum Genet. 2007; 15:103–109. PMID:
16969375.

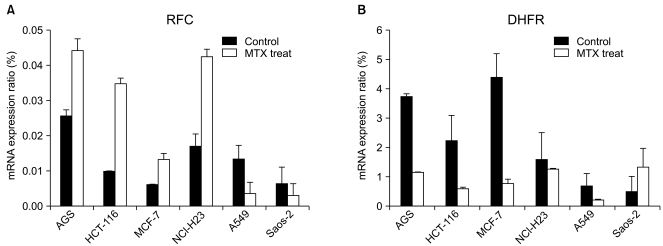
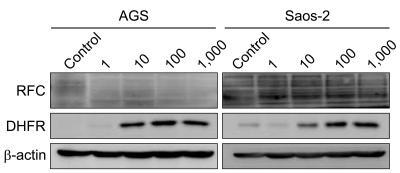
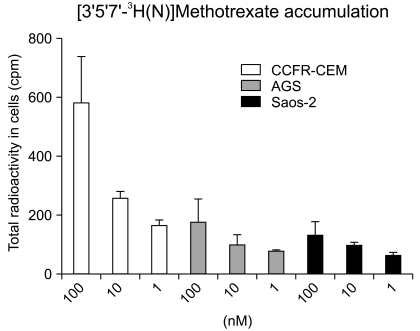
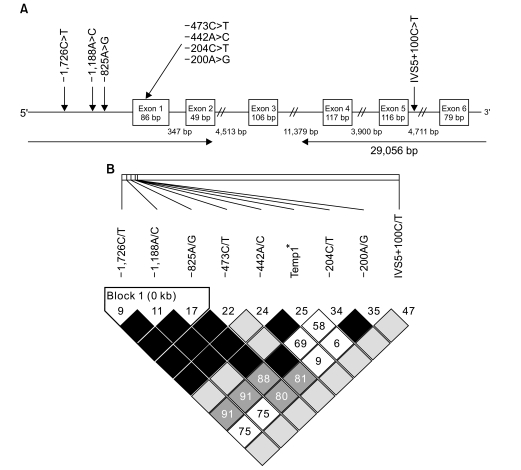




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download