Abstract
Purpose
Serine-threonine kinase11 (STK11) was originally identified in 1997 as the causative mutation that's responsible for Peutz-Jeghers Syndrome (PJS). Several recent studies have reported that the STK11 gene is an important human tumor suppressor gene in lung cancer. We evaluated the associations between the polymorphisms of the STK11 promoter region and the risk of lung cancer in 901 Koreans.
Materials and Methods
By direct sequencing, we first discovered three novel polymorphisms (-1,795 T>C, -981 C>T and -160 G>T) and four known polymorphisms (-1,580 C>T, -1,494 A>C, -881 A>G and -458 G>C) of the STK11 promoter region in 24 blood samples of 24 Korean lung cancer patients. Further genotype analyses were then performed on 443 lung cancer patients and 458 controls.
Results
We discovered three novel polymorphisms and we identified four known polymorphisms of the STK11 promoter region in a Korean population. Statistical analyses revealed that the genotypes and haplotypes in the STK11 gene were not significantly associated with the risk of lung cancer in a Korean population.
Conclusion
This is the first study that's focused on the association of STK11 promoter polymorphisms and the risk of lung cancer in a Korean population. To evaluate the role of the STK11 gene for the risk of lung cancer, the genotypes of the STK11 promoter region (-1,795 T>C, -1,494 A>C and -160 G>T) were determined in 901 Koreans, yet the result revealed no significant difference between the lung cancer patients and the controls. These results suggest that the three promoter polymorphisms we studied are not important risk factors for the susceptibility to lung cancer in Koreans.
The serine-threonine kinase11 (STK11) gene is a tumor suppressor gene that's known to be involved in several cellular processes, including signal transduction, energy sensing and cell polarity (1,2). Germ line mutations in the STK11 gene have been reported to be associated with Peutz-Jeghers polyposis and cancer syndrome (3,4). Somatic inactivating mutations in the STK11 gene have been shown in breast cancer, melanoma, colon cancer and lung cancer (5-9).
Several lines of evidences suggest that the STK11 gene is an important human tumor suppressor gene (10). Somatic STK11 alterations are rare in sporadic cancer with the exception of lung adenocarcinoma (11). The studies that have been conducted to detect somatic STK11 inactivation have strongly suggested that it has a role in human non-small cell lung cancer (NSCLC) (12,13). Approximately 30% of all somatic lung adenocarcinomas harbor an inactivating mutation (9), and a recent study has listed STK11 as one of the most frequently mutated tumor suppressor genes in lung adenocarcinoma (14).
Lung cancer has been the leading cause of cancer-related deaths in Korea, and its incidence continues to rise (15). Nevertheless, the prognosis of lung cancer has remained poor despite the innovations in diagnostic testing and surgical technique and the development of new chemotherapeutic agents. The recently introduced targeted agents show a different response according to the histologic subtype, and the efficiency of the treatment modalities for lung cancer depends on the time of diagnosis (16). Therefore, there is a great need for rapid and efficient methods for lung cancer's early detection. For developing improved molecular biomarkers for the early detection and prediction of a response to chemotherapy, it is important to identify the genetic alterations that are specific to each subtype of lung cancer. Single nucleotide polymorphisms (SNPs) have an excellent potential to be used as biomarkers for diagnosing genetic diseases, including cancers, compared to the other less common polymorphisms and microsatellite markers.
This study was conducted to examine the polymorphisms of the STK11 promoter region and their relation with the risk of lung cancer in a Korean population. We report here on the first identified polymorphisms of the STK11 promoter region in a Korean population of lung cancer patients.
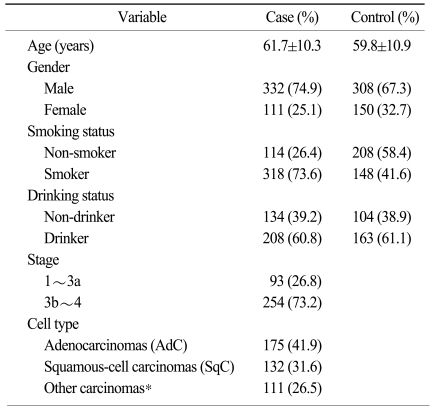
Between August 2001 and November 2007, blood samples were collected from 901 subjects, including 443 lung cancer patients and 458 normal controls without cancer. The lung cancer patients were recruited from the patient pool at the Genomic Research Center for Lung and Breast/Ovarian Cancer and the Inha University Medical Center, and the control subjects were randomly selected from a pool of healthy volunteers who had visited the Cardiovascular Genome Center, the Genomic Research Center for Allergy and Respiratory Diseases and the Keimyung University Dongsan Medical Center. Detailed information on diet, the smoking status, the drinking status, lifestyle and the medical history was collected by a trained interviewer with using a structured questionnaire. Out of 443 cases, 432 smoking statuses, 342 drinking statuses, 347 stages and 418 cell types were available for the information on the subjects' characteristics, while 356 smoking statuses and 267 drinking statuses, out of the 458 controls, were available for the information on the controls' characteristics. All the study subjects provided written consent and they were all ethnic Koreans, and all the participating Institutional Review Board approved the study protocol.
Genomic DNA was prepared from the peripheral blood samples using a Puregene blood DNA kit (Gentra, Minneapolis, MN) and following the manufacture's protocol.
For identification of the frequent polymorphism sites in the STK11 gene promoter in this Korean population, human genomic DNA was isolated from the whole blood of 24 samples for direct sequencing. Of the entire STK11 gene at 19p13, we amplified 2 kb of its promoter. PCR amplifications were performed in a PTC-225 Peltier Thermal cycler (MJ Research Inc, Waltham, MA) and with using AmpliTagGold (Roche, Branchburg, NJ). All the amplifications were performed using 35 cycles of 30 sec each cycle at 95℃, 1 min at 64℃ and 1 min at 72℃, and this was followed by a single 10 min extension at 72℃. The PCR products were purified using a Montage PCR96 Cleanup kit (Millipore, Bedfore, MA) and they were eluted in 20 µL of nuclease free H2O. DNA cycle sequencing was carried out using a BigDye Terminator V 3.1 Cycle Sequencing kit (Perkin Elmer, Foster City, CA). Multiscreen SEQ 384 well filter plates were used for dye terminator removal, and the sequences were analyzed on an Applied Biosystems 3700 DNA analyzer. All the polymorphisms and sequence alignments were analyzed using Polyphred.
After direct sequencing of the STK11 gene, we performed genotyping for the three polymorphisms (-1,795 T>C, -1,494 A>C and -160 G>T). The genotypes of the sample were assayed using a single base primer extension assay and employing a SNaPShot assay kit (ABI, Foster City, CA). Briefly, the genomic regions containing the polymorphisms of interest in the STK11 gene were amplified by PCR. The PCRs were performed using an initial denaturation step at 95℃ for 10 min, 35 amplification cycles (30 sec at 95℃, 1 min annealing at 63.9℃ and 1 min at 72℃), and this was followed by a single 7 min extension cycle at 72℃. The PCR products were subsequently purified by incubating them with 10 units of Exo I (USB, Cleveland, OH) and 1 unit of shrimp alkaline phosphatase (Roche, Indianapolis, IN) at 37℃ for 1 hour and then at 72℃ for 15 min. The extension reactions with 1 µL of purified PCR product, 0.15 pmoles of genotyping primer and a SNaPshot Multiplex Ready Reaction Mix (Applied Biosystems, Foster City, CA) were carried out by repeating the following cycle 25 times: 96℃ for 10 sec, 50℃ for 5 sec and 60℃ for 30 sec. The extension products were incubated with 1 unit of shrimp alkaline phosphatase (Roche, Indianapolis, IN) at 37℃ for 1 hour and then they were incubated at 72℃ for 15 min. 9 µL of deionized formamide was mixed with 1 µL of the purified extension product, and this was electrophoresed on an ABI Prism 3,700 genetic analyzer (Applied Biosystems, Foster City, CA). The results were analyzed using GeneScan analysis, version 3.7 (Applied Biosystems, Foster City, CA).
The allele frequencies, the genotype frequencies and the departures of the genotype distributions from Hardy-Weinberg equilibrium for each SNP were analyzed using the chi-square test or Fisher's exact test. A p-value of <0.05 was considered statistically significant. Linkage disequilibrium (LD) was tested on pairwise combinations of polymorphisms with using the absolute value of the standardized measure of linkage disequilibrium, D', as calculated by the Haploview program version 3.2. The haplotypes and their frequencies were estimated by the Haploview program version 3.2. The genotype-specific risks were estimated as odds ratios (ORs) with associated 95% confidence intervals by unconditional logistic regression analysis (SAS Institute, Cary, NC) and they were adjusted for age, gender and the smoking status.
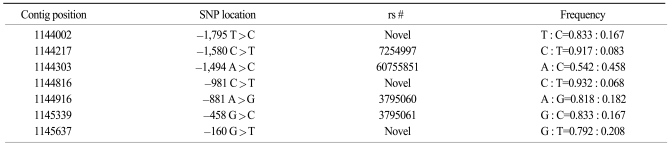
Table 1 shows the clinicopathological features of the cases and controls. By direct sequencing of the STK11 promoter region (~2 Kb), we discovered three novel polymorphisms and we identified four known polymorphisms of the STK11 promoter region in 24 lung cancer patient samples (Table 2). Among the seven SNPs, three SNPs (-1,975 T>C, -1,494 A>C and -160 G>T) were selected for large-scale genotyping based on their frequencies and LD status. Further analyses were then performed on the samples from 443 lung cancer patients and 458 controls. The genotype distributions of the polymorphisms among the population were in Hardy-Weinberg equilibrium.
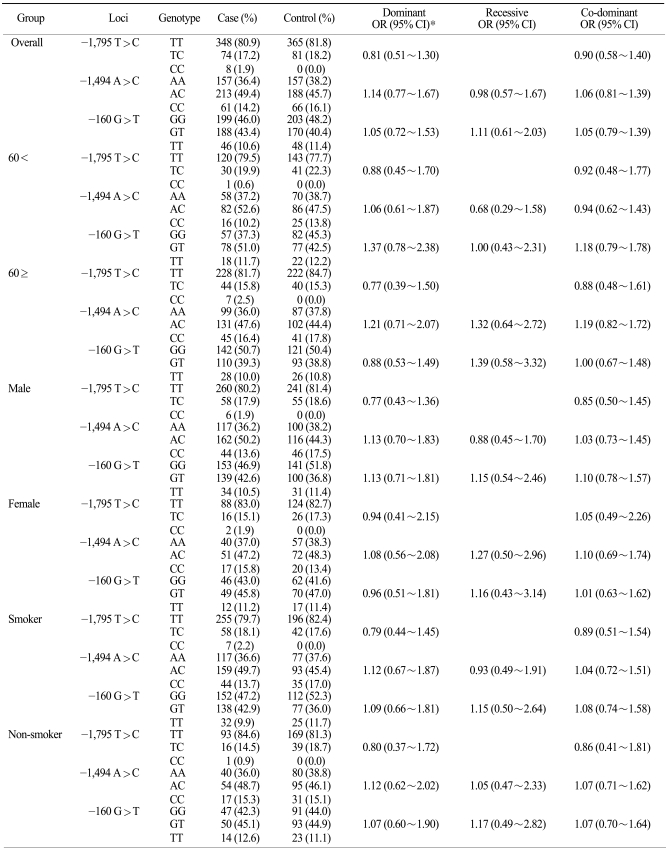
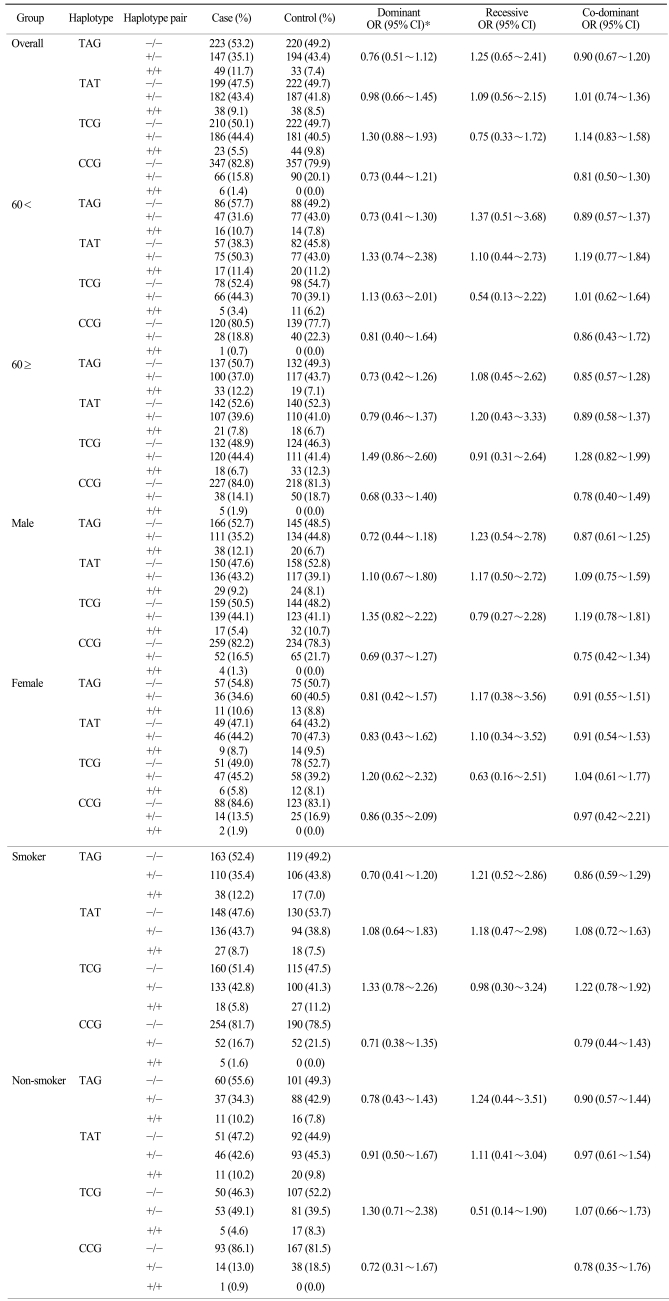
The association of the lung cancer risk with the STK11 promoter polymorphisms was then analyzed (Table 3). The subsequent analysis revealed that the -1,975 T>C polymorphism was not associated with the risk of lung cancer in dominant (OR: 0.81, 95% CI: 0.51~1.30) and co-dominant (OR: 0.90, 95% CI: 0.58~1.40) model. The -1,494 A>C polymorphism's association with the risk of lung cancer was not significant (dominant OR: 1.14, 95% CI: 0.77~1.67, recessive OR: 0.98, 95% CI: 0.57~1.67 and co-dominant OR: 1.06, 95% CI: 0.81~1.39). Also, the 160 G>T polymorphism's association with the risk of lung cancer was not significant (dominant OR: 1.05, 95% CI: 0.72~1.53, recessive OR: 1.11, 95% CI: 0.61~2.03 and co-dominant OR: 1.05, 95% CI: 0.79~1.39). The association of the polymorphisms with the risk of lung cancer was further examined after stratifying the subjects according to age, gender and smoking status. However, the subsequent analysis revealed no significant association. Furthermore, the haplotypes of the STK11 promoter polymorphisms were not associated with the risks of lung cancer in the three alternative models (Table 4).
In this study, a question of whether polymorphisms of the STK11 tumor suppressor gene are involved in the carcinogenesis of NSCLC was investigated by direct sequencing the promoter region of the STK11 gene of 24 Korean lung cancer patients. The result showed that the STK11 promoter polymorphisms were not associated with the risk of lung cancer in Korean population.
The human STK11 gene is a tumor suppressor gene that encodes an approximately 48 kDa protein of 436 amino acids with a serine-threonine kinase domain (17). The overexpression of STK11 protein leads to cell growth inhibition by G1 cell-cycle arrest (18), and this is associated with the Brahma protein homolog 1 (BRG1) and cellular tumor antigen p53 (19). Its involvement in the vascular endothelial growth factor (VEGFR) signaling pathway and in the phosphatase and tensin homolog (PTEN) pathway has also been demonstrated (20,21).
Somatic mutations were first described in 1998 in colorectal adenomas (22). Somatic mutations are relatively rare in sporadic cancers, and the only exception was shown by several studies to be lung adenocarcinomas (9,11). STK11 somatic mutations have been frequently detected in tumors from Caucasians, but not in the tumors of Asian lung cancer patients (23). An association between the polymorphism of genes encoding STK11 and the risk of type 2 diabetes has recently been reported (24), however, there has been no study on the polymorphisms of the STK11 promoter region in lung cancer. Polymorphisms often show ethnic variations (25). The polymorphisms of -1,795 T>C, -981 C>T and -160 G>T were polymorphisms of novel sites, and the polymorphisms of -1,580 C>T, -1,494 A>C, -881 A>G and -458 G>C are already known. However, the frequencies of these polymorphisms have not been reported in the dbSNP (www.nci.nlm.nih.gov/SNP) and HapMap database (www.hapmap.org). To the best of our knowledge, this is the first study on the association of the STK11 promoter polymorphisms and the risk of lung cancer in a Korean population.
The genotypes of the STK11 promoter region (-1,975 T>C, -1,494 A>C and -160 G>T) were determined in 901 Koreans to evaluate the role of the STK11 gene polymorphisms for the risk of lung cancer; however, the results revealed no significant difference between the lung cancer patients and the controls. This data suggests that polymorphisms in the STK11 promoter region are unlike to play an important role in the susceptibility to lung cancer.
Acknowledgement
We would like to thank Young Lim, Dong-Hoon Shin, Choon-sik Park and Yangsoo Jang for the sample collection and provision.
References
1. Spicer J, Ashworth A. LKB1 kinase: master and commander of metabolism and polarity. Curr Biol. 2004; 14:R383–R385. PMID: 15186763.

2. Mahoney CL, Choudhury B, Davies H, Edkins S, Greenman C, Haaften G, et al. LKB1/KRAS mutant lung cancers constitute a genetic subset of NSCLC with increased sensitivity to MAPK and mTOR signalling inhibition. Br J Cancer. 2009; 100:370–375. PMID: 19165201.

3. Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998; 391:184–187. PMID: 9428765.

4. Hezel AF, Bardeesy N. LKB1; linking cell structure and tumor suppression. Oncogene. 2008; 27:6908–6919. PMID: 19029933.

5. Avizienyte E, Loukola A, Roth S, Hemminki A, Tarkkanen M, Salovaara R, et al. LKB1 somatic mutations in sporadic tumors. Am J Pathol. 1999; 154:677–681. PMID: 10079245.

6. Bignell GR, Barfoot R, Seal S, Collins N, Warren W, Stratton MR. Low frequency of somatic mutations in the LKB1/Peutz-Jeghers syndrome gene in sporadic breast cancer. Cancer Res. 1998; 58:1384–1386. PMID: 9537235.
7. Forster LF, Defres S, Goudie DR, Baty DU, Carey FA. An investigation of the Peutz-Jeghers gene (LKB1) in sporadic breast and colon cancers. J Clin Pathol. 2000; 53:791–793. PMID: 11064676.

8. Guldberg P, thor Straten P, Ahrenkiel V, Seremet T, Kirkin AF, Zeuthen J. Somatic mutation of the Peutz-Jeghers syndrome gene, LKB1/STK11, in malignant melanoma. Oncogene. 1999; 18:1777–1780. PMID: 10208439.

9. Sanchez-Cespedes M, Parrella P, Esteller M, Nomoto S, Trink B, Engles JM, et al. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 2002; 62:3659–3662. PMID: 12097271.
10. Alessi DR, Sakamoto K, Bayascas JR. LKB1-dependent signaling pathways. Annu Rev Biochem. 2006; 75:137–163. PMID: 16756488.

11. Strazisar M, Mlakar V, Rott T, Glavac D. Somatic alterations of the serine/threonine kinase LKB1 gene in squamous cell (SCC) and large cell (LCC) lung carcinoma. cancer Invest. 2009; 27:407–416. PMID: 19229701.
12. Shah U, Sharpless NE, Hayes DN. LKB1 and lung cancer: more than the usual suspects. Cancer Res. 2008; 68:3562–3565. PMID: 18483235.
13. Makowski L, Hayes DN. Role of LKB1 in lung cancer development. Br J Cancer. 2008; 99:683–688. PMID: 18728656.

14. Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008; 455:1069–1075. PMID: 18948947.
15. Shin HR, Jung KW, Won YJ, Kong HJ, Yim SH, Sung J, et al. National cancer incidence for the year 2002 in Korea. Cancer Res Treat. 2007; 39:139–149. PMID: 19746208.

16. Brambilla E, Travis WD, Colby TV, Corrin B, Shimosato Y. The new World Health Organization classification of lung tumours. Eur Respir J. 2001; 18:1059–1068. PMID: 11829087.

17. Jenne DE, Reimann H, Nezu J, Friedel W, Loff S, Jeschke R, et al. Peutz-Jeghers syndrome is caused by mutations in a novel serine threonine kinase. Nat Genet. 1998; 18:38–43. PMID: 9425897.
18. Tiainen M, Ylikorkala A, Makela TP. Growth suppression by Lkb1 is mediated by a G(1) cell cycle arrest. Proc Natl Acad Sci U S A. 1999; 96:9248–9251. PMID: 10430928.

19. Karuman P, Gozani O, Odze RD, Zhou XC, Zhu H, Shaw R, et al. The Peutz-Jegher gene product LKB1 is a mediator of p53-dependent cell death. Mol Cell. 2001; 7:1307–1319. PMID: 11430832.

20. Ylikorkala A, Rossi DJ, Korsisaari N, Luukko K, Alitalo K, Henkemeyer M, et al. Vascular abnormalities and deregulation of VEGF in Lkb1-deficient mice. Science. 2001; 293:1323–1326. PMID: 11509733.
21. Jimenez AI, Fernandez P, Dominguez O, Dopazo A, Sanchez-Cespedes M. Growth and molecular profile of lung cancer cells expressing ectopic LKB1: down-regulation of the phosphatidylinositol 3'-phosphate kinase/PTEN pathway. Cancer Res. 2003; 63:1382–1388. PMID: 12649203.
22. Dong SM, Kim KM, Kim SY, Shin MS, Na EY, Lee SH, et al. Frequent somatic mutations in serine/threonine kinase 11/Peutz-Jeghers syndrome gene in left-sided colon cancer. Cancer Res. 1998; 58:3787–3790. PMID: 9731485.
23. Koivunen JP, Kim J, Lee J, Rogers AM, Park JO, Zhao X, et al. Mutations in the LKB1 tumour suppressor are frequently detected in tumours from Caucasian but not Asian lung cancer patients. Br J Cancer. 2008; 99:245–252. PMID: 18594528.

24. Keshavarz P, Inoue H, Nakamura N, Yoshikawa T, Tanahashi T, Itakura M. Single nucleotide polymorphisms in genes encoding LKB1 (STK11), TORC2 (CRTC2) and AMPK alpha2-subunit (PRKAA2) and risk of type 2 diabetes. Mol Genet Metab. 2008; 93:200–209. PMID: 17950019.
25. Sung JS, Han SG, Whang YM, Shin ES, Lee JW, Lee HJ, et al. Putative association of the single nucleotide polymorphisms in RASSF1A promoter with Korean lung cancer. Lung Cancer. 2008; 61:301–308. PMID: 18313166.





 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download