INTRODUCTION
MATERIALS AND METHODS
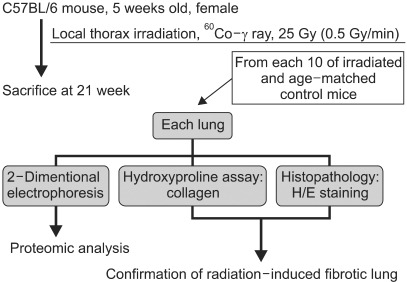
1) Induction of early phase of pulmonary fibrosis
2) Histological studies
3) Hydroxyproline assay
4) Sample preparation for 2-DE
5) Isoelectric focusing (IEF) and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)
6) In-gel digestion and MALDI-TOF analysis
7) Statistical analysis
RESULTS
1) Establishment of mouse RIF model
 | Fig. 2Confirmation of pulmonary fibrosis using H/E staining and the hydroxyproline assay. The mice were local thorax irradiated at a dose of 25 Gy (0.5 Gy/min). Twenty-one weeks later, severe collagen deposition and the initial phase of fibrosis developed. (A) Alveolar congestion, alveolar cell infiltration, and collagen lay-down can be seen in the 25 Gy irradiation slide (original magnification×40). (B) The increase in the level of collagen can be confirmed by the data (means±SD) representing two comparable experiments with four mice per group. |
2) Analysis of two-dimensional electrophoresis gel image
 | Fig. 32-DE of mouse lung. (A) representative proteome image of RIF. (B) normal lung tissue. (C) radiation-induced pulmonary fibrosis lung tissue. The proteins from the whole lung were extracted and separated on pH 3 to 10 linear immobilized pH-gradient strips, followed by a 10~16% sodium dodecyl sulfate/polyacrylamide gel. The gels were stained by silver staining. A total of 70 spots are tagged by numbers. (A) indicates the total up-regulated or down-regulated protein. The upper arrows indicate the protein spots corresponding to the α1-protease inhibitor, and the lower arrows indicate the protein spots corresponding to galectin-1. |
Table 1

The protein spots were excised and digested in a gel containing trypsin for protein identification using MALDI-TOF/MS. The MS spectra of the protein digests were searched against NCBInr and Swissprot, which are accessible at http://www.ncbi.nlm.nih.gov/ and http://www.expasy.com. *The scores are the confidence level of the match from the MASCOT program.
3) Up-regulation of α1-protease inhibitor and galectin-1 expression in RIF
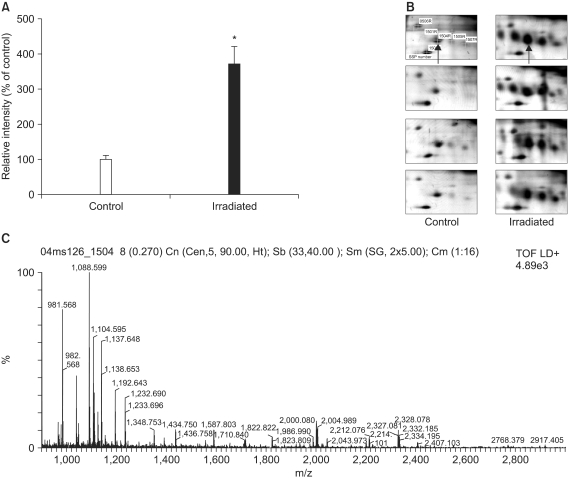 | Fig. 42-DE of murine α1-protease inhibitor. (A) Quantitation of the serine protease inhibitor in the normal lung tissues and RIF tissues. The protein spots that appeared in the 2DE gels were quantified using PDQuest computer software. The average intensities (n=8) of the spots corresponding to the α1-protease inhibitor in 2DE gels are shown. *p<0.005 compared to control. (B) 2DE gel images of α1-protease inhibitor. The arrow indicates the protein spot corresponding to the α1-protease inhibitor. (C) MALDI-TOF MS peptide mass spectrum of the tryptic digestion of the α1-protease inhibitor. |
 | Fig. 52-DE of murine galectin-1. (A) Quantitation of galectin-1 in the normal lung tissues and RIF tissues. The protein spots that appeared in the 2DE gels were quantified using PDQuest computer software. The average intensities (n=8) of the spots corresponding to galectin-1 in 2DE gels are shown. *p<0.005 compared to control. (B) 2DE gel images of galectin-1. The arrow indicates the protein spot corresponding to galectin-1. (C) MALDI-TOF MS peptide mass spectrum of the tryptic digestion of the gallectin-1. |




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download