INTRODUCTION
Cervical cancer is the second most common malignant tumor among women around the world. There are approximately 500,000 new cases of cervical cancer per year worldwide and there are approximately 200,000 deaths each year (
1).
The causal factor in cervical carcinogenesis is thought to be infection by the human papillomavirus (HPV). Approximately more than 90% of all cervical cancer cases are associated with HPV infection. Persistent viral infection with the strong, constitutive expression of the viral oncogenes
E6 and
E7 is a decisive step for malignant transformation (
2). There are more than one hundred known genotypes of HPV, but only some of them are associated with infection of the genital mucosa. The low risk types are associated with benign genital warts, and the high-risk types can cause cervical intraepithelial neoplasia (CIN). In the majority of cases, the HPV infections are clinically benign and they are cleared by the host immune system, but approximately 3% to 10% of women cannot clear their HPV infections. These women become HPV carriers and are at high risk for developing cervical cancer (
3). While HPV infection is known as one of the most important etiological factors to cervical cancer, other factors such as the usage of oral contraceptives, smoking, multiple pregnancies and coinfection with chlamydia and/or herpes simplex virus-2 may have a role (
4).
Cervical squamous cell carcinoma (SCC) has high risk of progression depending on the presence of such unfavorable prognostic factors as an advanced FIGO-stage and LN involvement. It has been known that the genes related to cellular proliferation, the association of the extracellular matrix, cellular adhesion molecules, proteases and cytokine are related to tumor progression. These findings are mainly based on the expression of genes at the transcriptional level (
5,
6).
Two-dimensional polyacrylamide gel electrophoresis (2-DE) has been used to examine the heterogeneity of protein expression in tissues from different tumors such as bladder (
7), breast (
8), colon-rectum (
9), lung (
10), and so on. The advantage of 2-DE is that the complex protein expression is analyzed qualitatively and quantitatively. 2-DE combined with MALDI-TOF-MS has been applied to identify cancer-specific protein markers. These markers can provide a basis for developing new methods for making early diagnosis and instituting effective treatment.
Intensive screening to search for the candidate genes/proteins in cervical cancer is needed, and especially to identify the genes/proteins that play a role in progression of this disease. Proteomic and genomic approaches are now on-going to identify tumor markers. Hellman and coworkers have reported the protein expression patterns in primary carcinoma of the vagina (
11).
In this study, we examined the differences of the protein expression profiles in human SCC and normal cervix tissues by using 2-DE and MALDI-TOF mass spectrometry. We then correlated these expression profiles with their possible different biological functions to identify the biomarkers and important pathways of cervical carcinogenesis.
Go to :

MATERIALS AND METHODS
1) Patient tissue samples
A total of 50 tissue biopsies were analyzed. 17 tissue biopsies from non-tumor cervix tissues and 33 tissue biopsies from SCC tissues were provided by St. Mary's Hospital of the Catholic Medical School, the Kyungpook Medical School Hospital and, Bucheon hospital of Soonchunhyang Medical School according to the procedures approved by the Institutional Review Board of the Catholic University. All samples were taken by experienced gynecologists and gynecological surgeons, and the samples were evaluated cytologically and histologically. Equal amounts of proteins from 17 non-tumor disease patient samples were mixed together to make the control samples. Of the 33 patients, 25 patients at the International Federation of Gynecology and Obstetrics (FIGO) stage I (IA1: 2, IA2: 1, IB1: 17, IB2: 5), 4 patients at stage II (IIA: 1, IIB: 3) and 4 patients at stage III (IIIA: 4) were included in this analysis. The mean age of the patients at the time of diagnosis was 48.9 years (range: 33~78 years).
2) Sample preparations
The SCC tissues were iodine tested before surgery. The regions of tumor incidence were excised and then divided for H&E staining and 2-DE analysis. The tissue biopsies for 2-DE were washed with 40 ml of buffer (50 mM Tris/HCl, pH 7.4) for 30 s and next snap frozen in liquid nitrogen. The frozen samples were crushed and finely mixed mechanically with using a mortar. Each sample was dissolved in five volumes of lysis buffer containing 8 M urea, 4% CHAPS, 0.5% Pharmalyte (pH 3~10), 100 mM DTT and DNase/RNase and protease inhibitors for 30 min at 4℃. After being sonicated three times for 10 s, the samples were centrifuged at 14,000 rpm for 30 min at 4℃. The supernatant was taken and the protein concentration was determined by the Bradford method.
3) Two-dimensional gel electrophoresis (2-DE)
For each sample, the equivalent of 250µg of solubilized protein that was dissolved in a 300µl volume of rehydration solution was loaded onto a 17 cm long IPG strip (Bio-Rad Laboratories, Hercules, CA). Isoelectric focusing was performed using a PROTEAN IEF CELL (Bio-Rad Laboratories) according to the protocol: 16 h of rehydration at 20℃, 2 h at 150 V, 2 h at 300 V, 2 h at 500 V, 1 h at 1,000 V, 2 h at 3,000 V and 40,000 Vhr at 8,000 V, giving a total of 48,900 Vhr. The current for each strip was limited to 50µA. After the IEF, each strip was immediately equilibrated with gentle shaking in 10 ml of an equilibration solution (50 mM Tris-HCl, pH 8.8) containing 6 M urea, 30% glycerol and 2% SDS for 15 min; this was done two times. Twenty mM DTT was added to the first equilibration step, and 2% (w/v) iodoacetamide was added to the second equilibration step. The second dimension gel electrophoresis was carried out on a 12% SDS-PAGE gel (1 mm gel thickness, 15 mA/gel) using a Protean II xi 2-D Cell (Bio-Rad Laboratories), and the proteins were visualized by Blum silver staining (
12). The stained gels were scanned using a CS 800 calibrated densitometer (Bio-Rad Laboratories). Spots detection, quantification and normalization were carried out using PDQuest 2-D software™ Version 7.2 (Bio-Rad Laboratories). 2D-PAGE was performed for each cancer sample and with using a control sample. The reproducibility of gel was about 80% or more, which reflected the small variances between the samples or the procedure runs. The normalization method was the total quantity in the analysis set.
4) In-gel protein digestion and MALDI-TOF mass spectrometry
The protein spots of interest were excised manually and then washed three times with mili Q-water. In-gel digestion for peptide mass fingerprinting (PMF) analysis was carried out. The excised spots were destained with destaining solution (15 mM potassium ferricyanide and 50 mM sodium thiosulfate) and next washed with 25 mM ammonium bicarbonate/50% acetonitrile until the gels became opaque and colorless. After drying with a vacuum concentrator (SpeedVac Plus, Holbrook, NY) the gel was rehydrated with 3~10µl of trypsin solution (10 ng/µl) at 4℃ for 30 min, and then it was incubated overnight at 37℃. The tryptic peptides were extracted with 60% acetonitrile and 0.1% trifluoroacetic acid, and then they were dried with a vacuum concentrator.
The tryptic peptides mixtures were dissolved in 0.5% trifluoroacetic acid (TFA). Using a saturated solution of α-cyano-4-hydroxycinnamic acid in 0.1% TFA/50% acetonitrile as a matrix, mass spectrometric analyses were performed using a MALDI R (Micromass, UK). Internal calibration of the MALDI mass data was applied using the masses of the two trypsin autolysis products: [M+H]
+ 842.51 and [M+H]
+ 2211.10 Da. The generated data was screened in databases with using a mass tolerance <50 ppm and the possible missed cleavage of trypsin was set as 1. The proteins with more than 4 matched peptides were thought to be significant. The Mascot program (
http://www.matrixscience.com) was used for searching the databases Swiss-prot or NCBInr.
5) Western blot analysis
Whole tissue biopsies were dissolved in lysis buffer containing 8 M urea, 4% CHAPS, 0.5% Pharmalyte (pH 3~10), 100 mM DTT, DNase/RNase and protease inhibitors. The protein concentration was evaluated by using the BioRad Protein-Assay kit (Bio-Rad Laboratories). 40µg of each protein sample was analyzed via SDS-PAGE and then it was transferred to the 10 Hybond-ECL membrane (Amersham, Sweden) at 250 mA for 1 h. The blotted membrane was blocked with 5% skim milk and next reacted with a primary antibody (SCCA-1, SCCA-2, Hsp27, Tropomyosin; Santa Cruz Biotechnology Inc., Santa Cruze, CA,, Annexin I; Pharmingen, San Jose, CA) at a concentration of 2µg/ml for 4 h at room temperature. After washing with Tris-buffered saline containing 0.1% Tween 20, the membrane was incubated with the horseradish peroxidase-conjugated secondary antibody at a dilution of 1:5,000. The protein bands were visualized using an ECL Kit (Amersham, Sweden) according to the manufacturer's protocol.
Go to :

RESULTS
The tissue samples from 17 patients with non-tumor disease and the tissue samples from 33 patients with SCC were analyzed by 2-DE and then silver stained. The histology of the patients was SCC and the mean patient age was 48.9 (SD: 11.6) years. The number of patients at each tumor stage was 25 patients at stage I, 4 at stage II and 4 patients at stage III. The high risk factors such as HPV infection (15 out of 33), lymph node involvement (9 out of 33) and lymph vascular space invasion (15 out of 33) were recorded. The clinical and histopathological characteristics of the tissues are listed in
Table 1.
Table 1
Clinical and histopathological characteristics of 33 patients

The down-regulated or up-regulated protein expressions between the normal cervix samples and the cervical SCC samples were evaluated using PDQuest 2-D software™. By making comparison with using PDQuest 2-D software quantification, the spots that showed a statistically meaningful expression difference (p-value<0.05) were selected or the spots that showed more than a 50% increase or decrease of intensity were defined as being the up- or down-regulated spots. According to this definition, among the 35 candidate proteins, 17 proteins were up-regulated and 18 proteins were down- regulated in the SCC tissues (
Fig. 1). The protein spots were excised from the gels, digested with trypsin and analyzed with using a MALDI-TOF mass spectrometer. The peptide mass fingerprints from the proteins were obtained and the resulting spectra were used to identify the proteins with employing the Mascot search program.
Fig. 2 shows the zoom images and the PDQuest 2-D quantifications of three identified proteins: 14-3-3ε, annexin 1 and squamous cell carcinoma antigen 2 (SCCA-2). The mass spectrum of one of the identified proteins, i.e., the tropomyosin beta chain and the sequences of the assigned peptides are presented in
Fig. 3. The Mascot score was 175, the number of matched peptides (including any missing cleavage) was 18 and the sequence coverage was 47%, which supports the fidelity of assignment. The up- or down- regulated proteins are listed in
Tables 2 and
3.
 | Fig. 1Comparison of the proteome by performing two-dimensional gel electrophoresis on normal tissues and cervical SCC tissues. Representative examples of the 2-DE gels derived from normal cervix tissue and from the cervical SCC tissue. The normal cervix (A) and cervical SCC (B) total proteins were separated by 2-DE with using IPG strips (pH 3~10) in the first dimension and 12% SDS-PAGE in the second dimension. The identified protein spots are indicated by numbers. The down-regulated (A) or up-regulated (B) proteins in the cervical SCC are indicated. 
|
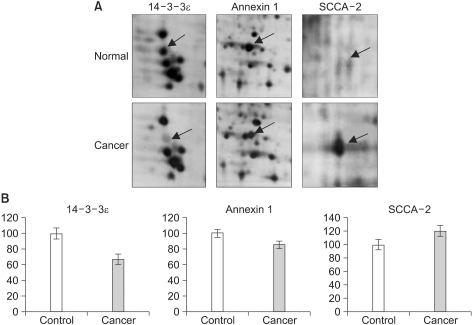 | Fig. 2Comparison of the protein expressions of the normal samples and the SCC samples. The up-regulated and down-regulated proteins (14-3-3ε, annexin A1 and SCCA-2) were selected and the magnified gel images are presented (A). The differences of expression were statistically meaningful according to the PDQuest 2-D software quantification (p-value <0.05) (B). 
|
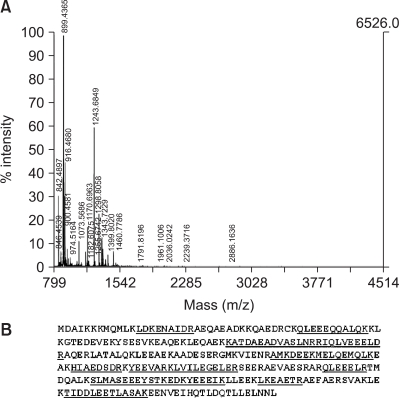 | Fig. 3MALDI-TOF peptide mass fingerprint of the tryptic digest of the tropomyosin beta chain. The MALDI-TOF mass spectrum of tropomyosin beta chain (A) and the matched peptide sequences are underlined (B). 
|
Table 2
Identification of protein spots by mass spectrometry
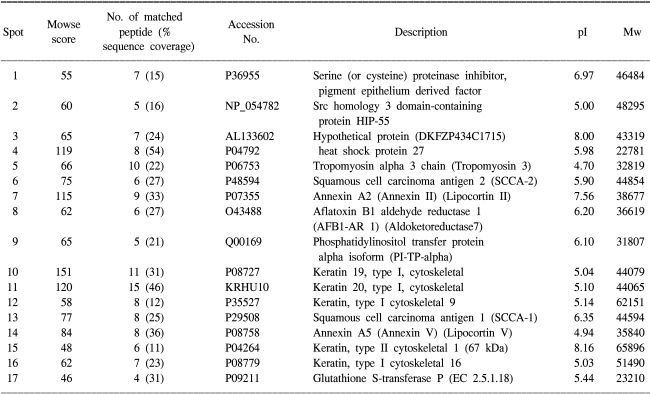

Table 3
Identification of protein spots by mass spectrometry
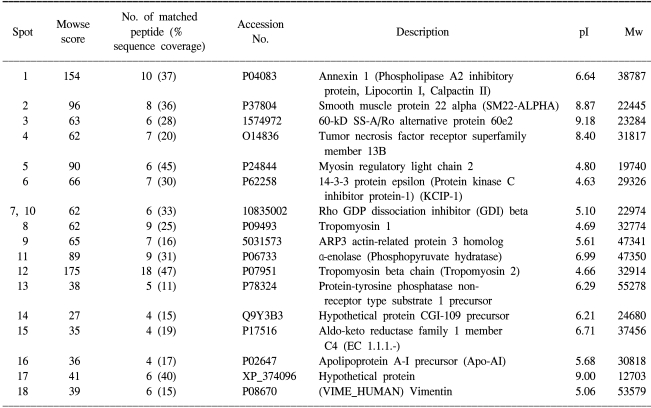

The 17 up-regulated proteins in the SCC tissues as compared to the normal tissues were keratin 1, 9, 13, 16, 19 and 20, aflatoxin B1 aldehyde reductase 1, annexin A2 and A5, heat shock protein 27, squamous cell carcinoma antigen 1 and 2, tropomyosin 3, serine (or cysteine) proteinase inhibitor, phosphatidylinositol transfer protein alpha isoform, Src homology 3 domain-containing protein HIP-55, glutathione S-transferase P and a hypothetical protein. The 18 down-regulated proteins in the SCC tissues were α-enolase, annexin 1, myosin regulatory light chain 2, 14-3-3ε, rho GDP dissociation inhibitor beta, ARP3 actin-related protein 3 homolog, tumor necrosis factor receptor superfamily member 13B, smooth muscle protein 22-alpha, tropomyosin 1 and 2, and hypothetical proteins. Western blot was performed to further verify the up-regulation or down-regulation of the proteins identified on the 2-DE and MALDI-TOF analyses. All the tested proteins, SCCA-1, SCCA-2, Hsp27, annexin 1 and tropomyosin showed the same expression patterns on 2-DE (
Fig. 4). These results also strengthen the validation of the 2-DE analysis in this report.
 | Fig. 4Western blot analysis of several proteins. The proteins obtained from the normal cervix control samples (N) and the cervical squamous cell carcinoma samples (C) were subjected to western blot analysis. Equal amounts of proteins (40 µg) were analyzed with SDS-PAGE and then they were Western blotted with anti-SCCA-1, anti-SCCA-2, anti-Hsp27, anti-tropomyosin and anti-annexin 1 antibodies. 
|
Although the high risk factors such as HPV infection (15 out of 33), lymph node involvement (9 out of 33) and lymph vascular space invasion (15 out of 33) were included in the study, there were no statistical significances between the high risk factors and the proteins that were differentially expressed. Also, there was no statistical tendency for the proteins expression in accordance with an early or late disease stage (data not shown).
Go to :

DISCUSSION
Using 2-DE and silver staining, we obtained the protein profiles of the normal cervix and the cervical SCC tissues. By comparing these profiles, we obtained 35 proteins that showed differential expressions in the normal and SCC tissues. The potential SCC-related proteins that were found through 2D gel proteomic analysis were proven to be limited because other candidate protein spots that had higher than 10 or lower than 5 pH values and lower detection limits for the determination of their masses could be easily excluded in this two-dimensional gel analysis. In this study, seventeen proteins were up-regulated in the SCC tissues, while 18 proteins were down-regulated (
Tables 2 and
3). Among the identified proteins, 12 proteins (pigment epithelium derived factor, annexin A2 and A5, keratin 19 and 20, heat shock protein 27, smooth muscle protein 22 alpha, α-enolase, squamous cell carcinoma antigen 1 and 2, glutathione S-transferase, and apolipoprotein a1) were previously known as proteins that are involved in tumor progression or they were differentially expressed by tumor, while 21 proteins were newly identified in this study.
Annexin A2 (ANXA2) is known as a Ca
2+ and phospholipid-binding protein, and the expression of ANXA2 is induced in various transformed cells (
13). Up-regulated ANXA2 has also been reported in human hepatocellular carcinoma, pancreatic adenocarcinoma, high-grade glioma, gastric carcinoma, acute promyelocytic leukemia and primary colorectal cancer. Like as been seen in other types of tumors, the ANXA2 expression was also up-regulated in this study. Since overexpression of the ANXA2 gene is commonly observed both in virally transformed cell lines and human tumors, it has been postulated that this up-regulated level of ANXA2 may link ANXA2 to a key step in cellular transformation and tumorigenesis. While the annexin A5 (ANXA5) expression is known to be elevated in cutaneous SCC and vaginal cancer, its physiological significance is not known. It has been known that the overexpression of annexin A1 (ANXA1) mediates the disruption of normal cell morphology and it inhibits the cyclin D1 expression and so reduces cell proliferation through proximal modulation of the ERK signal transduction pathway (
14). From our results, the expression of ANXA5 was up-regulated, while ANXA1 was down-regulated, and this implies different roles for these annexins in cervical SCC during tumorigenesis.
14-3-3 proteins are known as phosphoserine-binding protein (
15). The 14-3-3 proteins comprise a large family of highly conserved, small, acidic polypeptides that are 28-33 kDa in size, and they are found in all eukaryotic species. Among the isoforms of the 14-3-3 genes, 14-3-3σ has been most directly linked to cancer and it functions as a tumor suppressor by inhibiting cell-cycle progression and by causing cells to leave the stem-cell compartment and then undergo differentiation. Little was known about the cellular function of 14-3-3ε, yet 14-3-3ε is required for the correct timing of mitosis in
Drosophila and it is involved in cell-cycle progression in small cell lung cancer. 14-3-3εdown-regulation was observed in this study, which potentiates its role in SCC.
Heat-shock proteins (HSPs) promote cell survival under adverse environmental conditions. Synthesis of the 27-kDa, 70-kDa and 90-kDa HSPs is increased in malignantly transformed cells and this has been associated with tumor proliferation, metastasis and resistance to chemotherapeutic agents (
16). The increased expression of the 27-kDa heat-shock protein (HSP27) has been identified in human ovarian and mammary tumors. In patients suffering with cervical cancer, Hsp27 is predominantly expressed in well-differentiated SCCs and also moderately differentiated SCCs. Including our results, HSP 27 has recently been shown to be up-regulated in primary carcinoma of the vagina (
11), and in C33A cells that were transfected with the HPV
E7 gene (
17).
Down-regulation of α-enolase was reported in 26% of the cases of primary non-small cell lung cancer (NSCLC). Such down-regulation was associated with a poor clinical outcome, suggesting that α-enolase plays an important role in NSCLC. HPVE7 transfected C33A cells exhibited the elevated expression of α-enolase (
17). Further research on the expression of α-enolase in SCC is required.
Keratin was known as a biological marker in epithelial tissue differentiation (
18) and the cytoskeletal system is thought to be closely related to the malignant phenotype. An elevated cytokeratin-19 level has been associated with apoptotic resistance and the malignant progression of cervical carcinoma. The amplified product of cytokeratin 20 was found in the blood samples and in the fine-needle aspiration (FNA) biopsies of patients with thyroid carcinomas. In this study, the expressions of cytokeratin 19, 20, 16 and 1 were up-regulated in SCC. This cytokeratin expression may come about due to the cervical epithelial tissue's abnormal differentiation and uncontrolled proliferation.
Tropomyosins (TMs), myosin regulatory light chain and ARP3 actin-related protein are proteins related to the cellular cytoskeleton. It has been reported that the expression levels of TM1, TM2 and TM3 were reduced in T24 human bladder cancer cells and the overall changes in the TM expression levels seem to be an early event during bladder carcinogenesis (
19). The TM1 expression is known to be abolished in primary human breast tumors. TM1 also induced anoikis (detachment-induced apoptosis) in breast cancer cells, and the down regulation of TM1 in breast tumors may destabilize the microfilament architecture and confer resistance to anoikis, which would facilitate survival of neoplastic cells outside the normal micro-environment and so promote malignant growth. The alterations in the expression of the TM isoforms may provide further insight into the malignant transformation of SCCs of the cervix; this may be a useful target for new creating strategies for the early detection of SCC.
The connection between myosin regulatory light chain (MYL) 2, ARP3 actin-related protein and cervical tumorigenesis is not yet known. Yet the same as for tropomyosin, these proteins may participate in cell differentiation and the reorganization of the cytoskeleton during tumorigenesis. Further research on these proteins in cervical cancer will elucidate their possible role as tumor markers.
Smooth muscle protein 22 alpha (SM22-α) gene product is exclusively expressed in smooth muscle-containing tissues of adult animals and it is one of the earliest markers of differentiated smooth muscle cells (
20). It has been reported that SM22-α protein was down-regulated in lung, colon and breast cancer, which implied that loss of the SM22-α gene expression may be an important early event in tumor progression and also a possible diagnostic marker for the development of breast and colon cancer. SM22-α may be appropriate as a diagnostic marker for cervical SCC.
Squamous cell carcinoma antigen (SCCA) serves as a serological marker for more advanced squamous cell tumors of the cervix, lung and oropharynx (
21). SCCA has a molecular weight of about 45 kDa and it exists as two isoforms, i.e., the acidic and neutral isoforms. The neutral isoform of SCCA has been detected in both malignant and normal epithelial cells. In contrast, the acidic isoform is present in tumor cells, and especially those located at the periphery of the tumor; it is present in the sera of cancer patients with well-differentiated SCC (
22,
23). The functional role of SCCA in both normal and malignant cells has not been elucidated. It has been recently reported that SCCA2 has the ability to inhibit tumor necrosis factor alpha (TNF-α)-induced apoptosis in HeLa cells (
24) and XXXXto suppress SCCA, which inhibits tumor growth together with the increased infiltration of natural killer cells (
25). Interestingly, up-regulation of SCCA1 and SCCA2 and the concomitant down-regulation of the tumor necrosis factor (TNF) receptor superfamily were observed in our results. This might reflect the previous reported results concerning the relation between SCCA and the TNF-related apoptosis pathway. The concomitant increase in SCCA and the decrease in TNF receptor may confer cancer cells an advantage for survival and proliferation.
The relation between phosphatidylinositol transfer protein alpha isoform (PI-TP-α) and cancer is not yet known. PI-TP and phosphatidylinositol (PtdIns) 3-kinase together exhibit a capacity to phosphorylate PtdIns to PtdIns-3-phosphate. The existence of PI-TP-containing multiprotein complexes has been proposed; these may be poised to respond to extracellular stimuli and to effectively channel PtdIns from PI-TP towards the kinases and phospholipases for generating signal transducing molecules and regulating vesicular trafficking.
With using Gene Ontology analysis (
www.genmapp.org), the specific functions assigned to these proteins were correlated with the expression patterns for establishing a powerful cervical carcinogenesis pathway. The results showed that global induction was present in the processes of the cell cycle, epidermal differentiation, protein biosynthesis, RNA metabolism and protein metabolism in squamous cell cervical carcinoma. In contrast, significant down-regulation was shown in the processes of muscle development and cell adhesion, and there was damaged DNA binding activity. It can be suggested that there is a decrease in the adhesive properties of HPV-16 positive cervical cancer tissues cells, as compared to normal cells. For instance, rho GDP dissociation inhibitor beta protein was downregulated, which is thought to mediate the attachment, migration and organization of cells into the tissues. We could see that the GO analysis is descriptive of putative cervical carcinogenesis, and this suggests that several cellular functions were cervical cancer-dependent. Its role in cervical carcinogenesis is being further investigated.
As stated previously, 12 proteins such as annexin A2, α-enolase, keratin, tropomyosin and squamous cell carcinoma antigen (SCCA) have been previously shown to be cancer markers, while other proteins, including src homology 3 domain-containing protein HIP-55, PI-TP-α, 60-kDa SS-A/Ro alternative proteins and rho GDP dissociation inhibitor beta protein were newly identified in this study as proteins that are involved in cervical SCC. Although we could not completely explain all the correlations between the expressed proteins and their roles in tumorigenesis, it might be possible to find tumor- specific markers among these differentially expressed proteins. Further functional analysis of these proteins will bring forth more information on their physiological roles in the pathogenesis of SCC tumor.
Go to :










 PDF
PDF Citation
Citation Print
Print




 XML Download
XML Download