Abstract
Recent research in molecular biology has identified a significant number of novel markers, which may have diagnostic, prognostic and therapeutic significance. High-throughput tissue array method facilitates the validation of novel markers by enabling the simultaneous analysis of hundreds or thousands of tissue specimens. Tissue array slides can be analyzed using techniques such as immunohistochemistry and in situ hybridization. In this review, we give a brief overview of tissue array method and its application to high throughput clinicopathologic research.
Newly available genomic technologies and approaches now enable us to accumulate genetic information at a rapid pace. The sequencing of the human genome revealed an unprecedented amount of information about genes, their structure and variation (1,2). Based on these data, high-density cDNA microarrays enable the simultaneous analysis of the expression levels of thousands of genes (3). Similarly, modern proteomic tools allow survey of hundreds or thousands of proteins at once (4). By these techniques, multiple novel markers have been identified, primarily at the gene level. However, there remains a significant lag period between the discovery and validation of a gene as a clinical useful marker of therapeutic responsiveness or prognosis. The investigations of multiple markers on multiple tissue specimens are too time-consuming and labor intensive process using conventional methodology. The tissue array method is a high throughput molecular biology technique to overcome these problems.
In 1986, Hector Battifora described a 'sausage' block method, in which 1mm thick 'rods' of tissue, obtained from different specimens, were wrapped carefully in a sheet of small intestine (5). They described a method of embedding 100 or more different tissue samples in a normal-sized paraffin block, the multitumor tissue block. The multitumor tissue blocks allow the simultaneous immunohistologic testing of numerous tissue samples on a single slide with one drop of antibody. In 1990, he reported an improved method of 'checkerboard' (6). Because the tissues are evenly distributed in a checkerboard arrangement, they can be readily identified by their position in the resulting sections. Although this technique conferred a significant advantage of simultaneously examining multiple tissue specimens under identical conditions, the inability to satisfactorily identify individual 'rods' limited any meaningful interpretation. By extension of an idea originally developed by Battifora, tissue array method was first reported by Kononen et al. in 1998 (7).
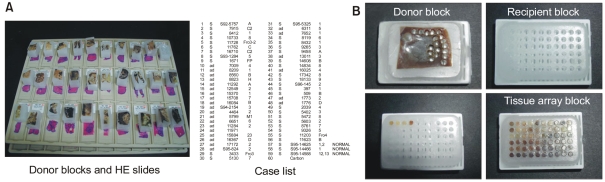
The first step in construction of a tissue array block is selection of cases from a database and collecting the respective hematoxylin and eosin (HE) stained slides from the archive. After reviewing all these slides, the area of study interest, commonly an area of cancer, is marked on the HE slides, in conjunction with an experienced histopathologist. The original histopathological blocks (the donor blocks) from the archive are collected and sorted with the marked slides. At the same time, the outline of the tissue array needs to be defined and a file should be generated that contains the identification numbers of the tissues together with their locations in array block (Fig. 1). After all of this preparatory work has been done, the tissue is ready to be arrayed and a tissue arraying device can be employed. Acquiring a tissue core from the donor block, and then this core is placed in precored hole in a recipient paraffin block using the arraying device. One of the tissue arraying system that is commercially available is Beecher Instruments (8). Using this manually operated device, excellent tissue array can be produced in the hands of a talented and experienced person. Although it is possible to take regular microtome sections from a tissue array block, a tape sectioning kit (Instrumedics Inc., St. Louis, MO) can facilitate slide cutting.
Using tissue array method, large-scale analyses of human tissues have been possible and consecutive sections from the array blocks allowed different protein expressions to be analyzed from defined, morphologically almost identical regions of the tumors. For example, if a tissue array block containing 1,000 cores is cut 200 times, as many as 200000 individual assays, and therefore outcomes can be produced from a single block (9). Schraml et al. (10) reported that a considerable fraction of the knowledge collected in about 100 previous investigations involving more than 8,000 experiments were reproduced by three experiments to a multitumor array, and during a 1-week period. Even in a large-scale analysis, tissue array method requires only small quantities of reagent and less laboratory personnel to perform the experiments. Tissue array method has proven to be extremely efficient and cost effective. Tissue array method also has the added advantage that all specimens are processed at one time using identical conditions including antigen retrieval, reagent concentration, incubation times with primary antibodies, and washing time. Slide conditions are also identical such as slide age or slide thickness. Furthermore, a tissue array block contains more than hundreds of specimens, and positive and negative controls are invariably included in a tissue array block. Therefore, tissue array methods can have an unprecedented level of standardization, over and above what is available using standard histopathological technique.
The histopathological benefits include minimal destruction of the original tissue blocks (11). Without considerable destruction of the original tissue blocks, researchers can infinitely store a few array blocks containing thousands of specimens as valuable resources, and can construct database and perform cohort study.
The most obvious question linked to tissue array method is to what extent tumor heterogeneity would affect the validity of the tissue array approach. The tissue size of a whole slide is about 2.5×2 cm, but the size of a small tissue array core is 0.6 mm, 1 mm or 2 mm in a diameter. A series of early studies using tissue array method showed that all findings that had previously been found by in situ methods on large sections, or by other methods on large tissue samples, could be fully reproduced in tissue array studies (7,10,11). And it could be concluded that the tissue array method is not analyzing the individual cases, but statistical analysis of a large number of cases.
Some previous studies providing evidence for the validity and representativity of tissue array data are summarized below. To assess these potential limitations of tissue array method, we constructed 4 array blocks (consisting of 60 cores with a diameter of 2 mm) from 4 different portions of each of 51 cases and the consistency of the staining results of MUC1 and MUC2 was evaluated by calculating the kappa values of Fleiss (12). The kappa value of MUC1 expressions was 0.74 (p<0.001) and that of MUC2 expressions was 0.87 (p<0.001). Therefore, an excellent agreement exists in the staining results of the intratumoral different areas of gastric carcinomas. Camp et al. (13) compared the staining of 2 to 10 array disks and the whole tissue sections from which they were derived. The results showed that even two cores of each tumor was comparable to analysis of a whole tissue section in more than 95% of cases, and that the degree of concordance increases to 99.5% with five cores per specimen. Hoos et al. (14) assessed the protein expression of Ki-67, p53, and pRb in full tissue sections of 59 fibroblastic tumors and compared with the expression status in one, two, and three 0.6 mm biopsies per tumor from the same specimens in an array. The results showed that the use of three cores per tumor gave optimal results, with concordance rates between tissue arrays with triplicate cores per tumor and full sections of 96, 98, and 91% for Ki-67, p53, and pRb staining, respectively.
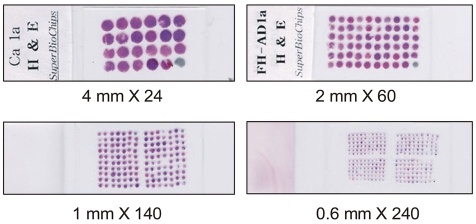
In the previous reports, the core with a diameter of 0.6 mm was frequently used (7,10,11,13,14). However, variable core size is possible such as 0.6 mm, 1 mm, 2 mm or 4 mm in a diameter (Fig. 2). In our opinion, a core with a diameter of 2 mm has some advantages. The core with a diameter of 2 mm contains ten-fold larger area for observation than the core with a diameter of 0.6 mm. Larger area enable the researchers to easily discriminate cancer cells from non-neoplastic cells. A tissue array block with 2 mm core contains up to 60 cores, and the researchers can satisfactorily identify the location of individual core in tissue arrays. Therefore, more accurate and easier assessment of tissue array slide is possible using 2 mm sized core. From a tissue array block with 2 mm sized core, we can take regular microtome sections, but sectioning of a tissue array block with 0.6 mm sized core require an adhesive tape technique. Some tissue cores are frequently lost during sectioning and immunostaining procedure. In our experiments, about 5% of tissue cores are lost for tissue array blocks with a diameter of 2 mm. However, about 20% of tissue cores are lost for tissue array blocks with a diameter of 0.6 mm irrespective of using an adhesive tape (15).
At the present time, 80% of tissue arrays produced is analyzed using immunohistochemistry, whereas most of the remaining tissue arrays are being investigated by in situ hybridization techniques, such as FISH (16). All research involving in situ tissue analysis can be done in tissue array format. For the in situ analysis, the construction of tissue arrays is flexible, meeting the focused needs of the investigator. Most of the previous applications of the technology were based on several different types of tissue arrays.
To develop prognostic markers for clinical applications, we constructed six array blocks containing 329 consecutive gastric carcinomas. About one hundred proteins have been evaluated immunohistochemically and 30 candidate prognostic markers including MUC1 (12), CD24 (17), MGMT (18), and smad7 (19) were related to patient outcome in gastric carcinomas. The tissue array method enabled us to analyze a large number of gastric carcinomas and consecutive sections from the array blocks allowed different protein expressions to be analyzed from defined, morphologically almost identical regions of the tumors. Therefore, the analysis of the combined status of different proteins and the analysis of each subgroup determined according to the pTNM stage were possible. Recently, expression loss of DNA-PKcs was reported to be significantly associated with poor survival, and moreover, loss of DNA-PKcs expression was identified to correlate with a lower survival in the subgroup of stage I (p=0.037) (20). It has been suggested that combination of expression of novel markers may be of more benefit in predicting prognosis. When we analyzed the relationship between the patient survival and the combined status of MUC1 or MUC2 and p53 protein expression, the patients with MUC1-/p53-pattern showed a better outcome than those with MUC1+/p53-, MUC1+/p53+ or MUC1-/p53+ expression patterns (p=0.001) (12). In contrast, the patients with MUC2-/p53+ pattern showed a worse outcome than those with MUC2+/p53+, MUC2+/p53- or MUC2-/p53- expression patterns (p=0.003).
Kang et al. (21) reported that 20% of gastric cancers showed expression loss of PTEN using the above tissue arrays. To compare protein expression status with DNA methylation status, the cases with expression loss of PTEN and the cases with intact PTEN expression were selected according to the results of tissue arrays. Expression loss of PTEN significantly correlated with promoter methylation of PTEN. Woo et al. (22) constructed three array blocks containing 162 gastric cancers and immunostaining against (-catenin was performed. A nuclear staining pattern was detected in 17.3% of gastric cancer and PCR-based sequencing of (-catenin exon 3 was performed for the nuclear staining cases and some of nuclear-negative cases. The mutation of (-catenin was detected in only 5% of gastric cancer. We could use the tissue array method as the primary screening for the genetic alteration study, even though the prevalence of genetic alterations has been predicted to be very low.
In our laboratory, 21 tissue array blocks, containing 1127 consecutive gastric cancers over a period of 2 years, were constructed (23). Sixty-three out of 1127 (5.6%) cases were Epstein-Barr virus (EBV)-positive gastric cancer by EBER in situ hybridization. To characterize the EBV-positive gastric cancer, an array block containing 63 EBV-positive cancers was constructed and evaluated by immunohistochemistry. EBV-positive gastric carcinomas were found to have a distinct protein expression profile in comparison with EBV-negative carcinomas. The composition of tissue arrays can be variable and flexible, and the researchers can make proper arrays for the focused needs of the studies.
Recently, some studies contain relatively large number of markers in addition to a large number of cases. We evaluated immunohistochemically the expression status of 20 tumor-associated proteins in 329 consecutive gastric cancers using tissue array method (24). In this study, the cumulative expression loss of tumor suppressor genes in gastric carcinoma has been suggested to be important in determining patient survival.
Using immunohistochemistry on tissue arrays, Jacquemier et al. (25) have monitored the expression of 26 selected proteins in more than 1,600 cancer samples from 552 consecutive patients with early breast cancer. Hierarchical clustering was performed and identified relevant clusters of coexpressed proteins and clusters of tumors. By clustering, subclasses of breast cancer could be identified and the subclasses predicted the metastasis free survival.
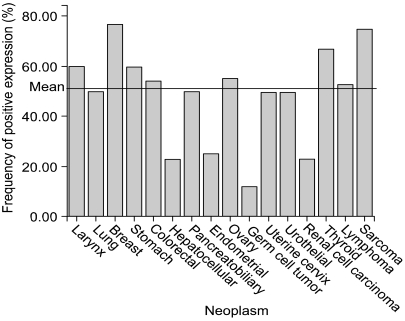
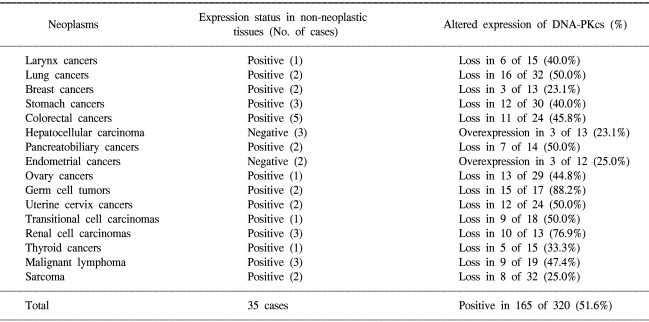
Whilst tumor arrays provide prognostic information to clinicians, arrays containing many tumor types can provide useful diagnostic information or different tumor biology according to tissue type. Multi-tumor arrays can provide the information of differential expression status of molecular markers in various organs. To clarify organ specificity of altered expression of DNA-PKcs, we constructed three tissue array blocks containing 320 cases of various malignant neoplasm and performed immunohistochemical staining against DNA-PKcs. Loss of DNA-PKcs expression was found in neoplasm originating from various organs with similar frequency, but germ cell tumors and renal cell carcinomas had significantly higher rate of loss of DNA-PKcs expression in comparison to other neoplasm (Table 1, Fig. 3) (unpublished data). Andersen et al. (26) performed a large-scale survey on the distribution and frequency of the 17q23 copy number increases across different tumor types using fluorescence in situ hybridization on tissue microarrays containing 4788 specimens, consisting 166 different tumor categories and 40 different tissue categories. Increased 17q23 copy number was detected in 15% of the evaluable specimens with tumors originating from the lung, mammary gland, and soft tissue being most frequently affected.
In addition to cancer specimen, precancerous lesions such as adenoma, dysplasia, and intraepithelial neoplasia were investigated using tissue array method. We constructed an array with 59 gastric adenomas and mucin expression status was evaluated (12). The rate of MUC1 expression in gastric carcinomas was significantly higher than in associated gastric adenomas (p<0.01).
Normal tissue arrays are especially important if candidate proteins are being evaluated for their potential utility as diagnostic reagents or therapeutic targets. The array blocks containing various normal and cancer tissue are useful for the expression screening or test of candidate genes. For this purpose, we constructed the human control slides for immunohistochemistry (Superbiochips Laboratories, Seoul, Korea). Control slides contain skin, breast, pancreas, lymph node, stomach, lung, salivary gland, liver, bile duct, spleen, gallbladder, stomach, small intestine, colorectum, kidney, prostate, seminal vesicle, testis, uterus, placenta, adrenal, thyroid, brain and their various tumors. For example, antibody against Hepatocyte was evaluated immunohistochemically using human control slide, and Hepatocyte was found to be expressed only in hepatocytes, hepatocellular carcinoma, intestinal metaplasia of stomach and in the mucosal columnar cells of small intestine (27). Based on these screening results, we could plan the further study, and immunostaining for hepatocyte was performed in many specimens of hepatocellular carcinoma, stomach cancer, intestinal metaplasia of gastric mucosa and fetal tissue of intestine. Many laboratories performed immunohistochemistry in daily practice and quality control in immunohistochemistry is one of the major problems. The test procedure using tissue array control slides may be helpful to the quality control.
A recently described use for tissue array methods includes the analysis of frozen tissue and cell lines (16). One difficulty of tissue arrays with paraffin-embedded tissue relates to antigenic changes in proteins and mRNA degradation induced by the fixation and embedding process. Fejzo et al. (28) described modified tissue array method using frozen tissues embedded in OCT compound as donor samples and arraying the specimens into a recipient OCT block. They showed OCT arrays work well for DNA, RNA, and protein analyses, and may have significant advantages for the assessment of some genes and proteins. However, many antibodies are now commercially available on formalin-fixed and paraffin-embedded tissues, and only a few subsequent studies using frozen tissues have been reported.
Attempts have been made to automate the process of tissue array methods, and major attempts are now in progress to automate the data analysis step using digital imaging and analyzing system (29). These automations will reduce the amount of manual work that is required. Improvements that are already increasing the overall efficiency of tissue arrays, such as the development of database structures that allow sharing of data and images across the Internet, are also contributing to its increased use.
Tissue array method has become a widely accepted standard technology. Tissue array method is valuable and efficient for the high throughput investigation of candidate genes and their proteins. Tissue array method plays an ever increasing role in translational research for a more rapid application of novel marker to clinical practice.
References
1. Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002; 420:520–562. PMID: 12466850.

2. Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001; 291:1304–1351. PMID: 11181995.

3. Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995; 270:467–470. PMID: 7569999.

4. Templin MF, Stoll D, Schrenk M, Traub PC, Vohringer CF, Joos TO. Protein microarray technology. Trends Biotechnol. 2002; 20:160–166. PMID: 11906748.

5. Battifora H. The multitumor (sausage) tissue block: novel method for immunohistochemical antibody testing. Lab Invest. 1986; 55:244–248. PMID: 3525985.
6. Battifora H, Mehta P. The checkerboard tissue block. An improved multitissue control block. Lab Invest. 1990; 63:722–724. PMID: 2232717.
7. Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998; 4:844–847. PMID: 9662379.

8. Fedor HL, De Marzo AM. Practical methods for tissue microarray construction. Methods Mol Med. 2005; 103:89–101. PMID: 15542899.

9. Mousses S, Kallioniemi A, Kauraniemi P, Elkahloun A, Kallioniemi OP. Clinical and functional target validation using tissue and cell microarrays. Curr Opin Chem Biol. 2002; 6:97–101. PMID: 11827831.

10. Schraml P, Kononen J, Bubendorf L, Moch H, Bissig H, Nocito A, et al. Tissue microarrays for gene amplification surveys in many different tumor types. Clin Cancer Res. 1999; 5:1966–1975. PMID: 10473073.
11. Bubendorf L, Nocito A, Moch H, Sauter G. Tissue microarray (TMA) technology: miniaturized pathology archives for high-throughput in situ studies. J Pathol. 2001; 195:72–79. PMID: 11568893.
12. Lee HS, Lee HK, Kim HS, Yang HK, Kim YI, Kim WH. MUC1, MUC2, MUC5AC, and MUC6 expressions in gastric carcinomas: their roles as prognostic indicators. Cancer. 2001; 92:1427–1434. PMID: 11745219.
13. Camp RL, Charette LA, Rimm DL. Validation of tissue microarray technology in breast carcinoma. Lab Invest. 2000; 80:1943–1949. PMID: 11140706.

14. Hoos A, Urist MJ, Stojadinovic A, Mastorides S, Dudas ME, Leung DH, et al. Validation of tissue microarrays for immunohistochemical profiling of cancer specimens using the example of human fibroblastic tumors. Am J Pathol. 2001; 158:1245–1251. PMID: 11290542.

15. Kim WH. Tissue array method for large scale clinicopathologic study. Korean J Pathol. 2002; 36:199–204.
16. Shergill IS, Shergill NK, Arya M, Patel HR. Tissue microarrays: a current medical research tool. Curr Med Res Opin. 2004; 20:707–712. PMID: 15140337.

17. Darwish NS, Kim MA, Chang MS, Lee HS, Lee BL, Kim YI, et al. Prognostic significance of CD24 expression in gastric carcinoma. Cancer Res Treat. 2004; 36:298–302.

18. Bae SI, Lee HS, Kim SH, Kim WH. Inactivation of O6-methylguanine-DNA methyltransferase by promoter CpG island hypermethylation in gastric cancer. Br J Cancer. 2002; 86:1888–1892. PMID: 12085181.
19. Kim YH, Lee HS, Lee HJ, Hur K, Kim WH, Bang YJ, et al. Prognostic significance of the expression of Smad4 and Smad7 in human gastric carcinomas. Ann Oncol. 2004; 15:574–580. PMID: 15033661.

20. Lee HS, Yang HK, Kim WH, Choe G. Loss of DNA-dependent protein kinase catalytic subunit (DNA-PKcs) expression in gastric cancers. Cancer Res Treat. 2005; 37:98–102.

21. Kang YH, Lee HS, Kim WH. Promoter methylation and silencing of PTEN in gastric carcinoma. Lab Invest. 2002; 82:285–291. PMID: 11896207.

22. Woo DK, Kim HS, Lee HS, Kang YH, Yang HK, Kim WH. Altered expression and mutation of beta-catenin gene in gastric carcinomas and cell lines. Int J Cancer. 2001; 95:108–113. PMID: 11241321.
23. Lee HS, Chang MS, Yang HK, Lee BL, Kim WH. Epstein-Barr virus-positive gastric carcinoma has a distinct protein expression profile in comparison with Epstein-Barr virus-negative carcinoma. Clin Cancer Res. 2004; 10:1698–1705. PMID: 15014022.

24. Lee HS, Lee HK, Kim HS, Yang HK, Kim WH. Tumor suppressor gene expression correlates with gastric cancer prognosis. J Pathol. 2003; 200:39–46. PMID: 12692839.
25. Jacquemier J, Ginestier C, Rougemont J, Bardou VJ, Charafe-Jauffret E, Geneix J, et al. Protein expression profiling identifies subclasses of breast cancer and predicts prognosis. Cancer Res. 2005; 65:767–779. PMID: 15705873.
26. Andersen CL, Monni O, Wagner U, Kononen J, Barlund M, Bucher C, et al. High-throughput copy number analysis of 17q23 in 3520 tissue specimens by fluorescence in situ hybridization to tissue microarrays. Am J Pathol. 2002; 161:73–79. PMID: 12107091.

27. Lee HS, Kim WH, Kang GH. Hepatocyte expressions in hepatocellular carcinomas, gastrointestinal neoplasms, and non-neoplastic gastrointestinal mucosa: its role as a diagnostic marker. J Korean Med Sci. 2003; 18:842–848. PMID: 14676441.

28. Fejzo MS, Slamon DJ. Frozen tumor tissue microarray technology for analysis of tumor RNA, DNA, and proteins. Am J Pathol. 2001; 159:1645–1650. PMID: 11696425.
29. Henshall S. Tissue microarrays. J Mammary Gland Biol Neoplasia. 2003; 8:347–358. PMID: 14973378.





 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download