Abstract
DNA microarray technology permits simultaneous analysis of thousands of DNA sequences for genomic research and diagnostics applications. Microarray technology represents the most recent and exciting advance in the application of hybridization-based technology for biological sciences analysis. This review focuses on the classification (oligonucleotide vs. cDNA) and application (mutation-genotyping vs. gene expression) of microarrays. Oligonucleotide microarrays can be used both in mutation-genotyping and gene expression analysis, while cDNA microarrays can only be used in gene expression analysis. We review microarray mutation analysis, including examining the use of three oligonucleotide microarrays developed in our laboratory to determine mutations in RET, β-catenin and K-ras genes. We also discuss the use of the Affymetrix GeneChip in mutation analysis. We review microarray gene expression analysis, including the classifying of such studies into four categories: class comparison, class prediction, class discovery and identification of biomarkers.
Microarray technology is a powerful platform for biological exploration (1). Microarrays permit simultaneous analysis of thousands of DNA sequences for genomic research and diagnostic applications. This technology represents the most recent and exciting advance in the application of hybridization-based approaches to analysis in the biological sciences (1). Gene expression profiling of cancers represents the largest research category using microarrays and appears to be the most robust approach for molecular characterization of cancers (2). It is becoming recognized that microarray technology will be a fundamental tool for future genomic research.
Depending on the type of probes used, microarray systems are classified as either oligonucleotide or cDNA. Oligonucleotide microarrays usually consist of a hybridization slide spotted with oligonucleotides ranging in length from 16-70-mer. Oligonucleotide microarrays can be used for gene expression, mutation, SNP (single nucleotide polymorphism) and genotyping analyses. Oligonucleotide microarrays have been developed as a method for rapid mutation analysis of selected gene sequences, and are effective in sequence analysis, diagnosing genetic diseases and gene polymorphism studies (3). A typical DNA microarray-based method is less time-consuming and is cheaper than conventional sequencing, and plays a valuable role in high throughput sequence analysis (4,5).
cDNA microarrays are usually used for analyzing gene expression. The expression of thousands of genes can be analyzed at the one time. Using cDNA microarrays is relatively easy and as such they are used in many research groups. A cDNA microarray usually consists of a slide spotted with cDNA probes ranging in size from a few hundred to 1,000 bp.
Microarray technology is usually used for gene expression profiling. The expression of tens of thousands of genes can be analyzed at one time. In addition to gene expression analysis, the microarray technique has been developed for mutation or SNP detection using allele-specific hybridization involving oligonucleotides. This type of microarray is called an 'oligonucleotide microarray' (6). Oligonucleotide microarrays can detect mutations or SNP by discriminating between perfectly matched and mismatched signals. Commercially available oligonucleotide microarrays include the p53 GeneChip and HuSNP arrays from Affymetrix, which were manufactured using photolithographic techniques (7). Several oligonucleotide microarrays have been developed for detecting K-ras mutations, methylation and RET mutations in MEN2 (Multiple Endocrine Neoplasia type 2) syndromes (8~10). Oligonucleotide microarrays show a high sensitivity in terms of point mutation detection (8), and can function as fast and reliable genetic devices, which simplifies detecting mutations.
Mutation analysis of genes has accelerated our understanding of disease-related mechanisms, which has had an impact in terms of both basic knowledge and clinical practice. However, each gene has different mutation characteristics and a different size. Most of the APC, BRCA1 and BRCA2 mutations found in cancers are protein truncating nonsense or frameshift mutations. However, missense mutations dominate in hot-spot areas for the RET proto-oncogene responsible for MEN2 syndromes (8). Several mutation detection techniques have been developed, such as PCR-SSCP (Polymerase Chain Reaction-Single Strand Conformational Polymorphism), PTT (Protein Truncation Test), PCR-RFLP (Polymerase Chain Reaction-Restriction Fragment Length Polymorphism), and sequencing. These conventional methods are widely used because they are easily performed and the experimental conditions are well known. The predominant RET mutations are missense mutations and are restricted to 10 codons (codons 609, 611, 618, 620, 630, 631, 634, 768, 804 and 918) in MEN2 syndromes. Missense mutations at codons 609, 611, 618, 620 and 634 have been identified in 98% of MEN2A families and in 85% of FMTC families. More than 95% of MEN2B patients also have a predominant mutation at codon 918 (Met→Thr). The RET oligonucleotide microarray can detect RET missense mutations at these 10 codons (8). Sixty-five oligonucleotides were designed for the 65 mutation types at 10 codons, and 12 oligonucleotides were designed for the wild-types and positive controls. The RET oligonucleotide microarray can function as a fast and reliable genetic diagnostic device, which simplifies the process of detecting RET mutations (Fig. 1).
Not only does β-catenin function as a downstream transcriptional activator in the Wnt signaling pathway, it is also a submembrane component of the cadherin-mediated cell-cell adhesion system (11,12). β-catenin mutations have been identified in a variety of human malignancies, with most of these being missense mutations restricted to hot-spot areas in exon 3. In many human cancers, including endometrial, gastric, ovarian, hepatoblastomas and colorectal cancers, the majority of β-catenin mutations have been reported at specific GSK-3 phosphorylation sites, i.e., Ser-33, Ser-37, Thr-41, Ser-45 and other residues (Asp-32 and Gly-34) (13). More than 70 β-catenin mutations have been reported in colorectal cancers, and about 90% of β-catenin mutations are found in 11 codons (codons 29, 31, 32, 33, 34, 35, 37, 38, 41, 45 and 48) as missense mutations or in-frame deletions. We have developed an oligonucleotide microarray for detecting β-catenin mutations at these 11 codons (14). This microarray can detect a total of 110 types of β-catenin mutation, including the previously reported 60 mutations. All oligonucleotides on that array were 21 bp long and the mismatch sequence was located in the middle of the oligonucleotide. Oligonucleotides were modified with 5'-amino residues for chemical binding to the slides. Twelve-carbon spacers were used to increase the efficiency of the hybridization and to make it easier for target samples (labeled with fluorescent dye) to access the binding site. One hundred and ten oligonucleotides were designed for codons 29, 31, 32, 33, 34, 35, 37, 38, 41, 45 and 48, and eleven oligonucleotides were designed to detect in-frame deletions in the eleven codons. In total, the 121 designed oligonucleotides covered all substitutions and in-frame deletions in the above eleven codons of exon 3.
Activating mutations of the K-ras gene occur in approximately 20~50% of colorectal cancers, with ~85% of the mutations restricted to codons 12 and 13 (15). Studies of associations between K-ras mutations and specific clinical features generally require analysis of large numbers of samples (16). Thus, researchers need a high-throughput technique for assessing K-ras mutations. Oligonucleotide microarrays may provide a valid option, as they allow scientists to accurately and rapidly process large numbers of samples. We developed a new method of K-ras oligonucleotide microarray analysis called Competitive DNA Hybridization (CDH). CDH is a novel, efficient, high capacity hybridization technique involving the mixing of various fluorescent-labeled samples to compete with each other in the hybridization reaction. For this work, we used dNTPs labeled with Cy5-dCTP, Cy3-dCTP and Alexa™ 594-dUTP, which have distinct spectra. Accordingly, our CDH results showed improved microarray imaging due to less non-specific hybridization.
An Affymetrix GeneChip™ is produced by synthesizing tens of thousands of short oligonucleotides in situ on glass wafers, one nucleotide at a time, using a modification of semiconductor photolithography (17). There are several differences between GeneChips and homemade cDNA microarrays or other oligonucleotide microarrays. In the GeneChip system, both gene expression arrays and variation detection arrays are available. About 4-5 variation detection arrays (p53, HIV, SNP, CYP450, BRCA1) have been reported as being used, and p53 arrays are used for p53 mutation detection by a wide variety of cancer research groups (18). The Affymetrix P450 GeneChip is used for pharmacogenetic screening. This GeneChip is an efficient and reliable tool for testing CYP2D6 gene variation based on five alleles (19). Developing oligonucleotide microarrays like the GeneChip arrays requires very high level synthesis and photolithography technologies.
Early use of photolithography techniques allowed the production of a GeneChip containing more than 65,000 different oligonucleotides in an area of 1.6 cm2 (20). Although originally developed for mutation detection, the same technology was adapted to measure expression levels of cytokine genes in murine T cells (21). In general, 11~16 probes are selected among all possible 25-mers to represent each transcript. In addition to choosing the probes based on their predicted hybridization properties, candidate sequences are filtered for specificity. A core element of the GeneChip design, the 'Perfect Match/Mismatch' probe strategy, is also universally applied to the production of GeneChip arrays. For each probe designed to be perfectly complementary to a target sequence, a partner probe is generated that is identical except for a single base mismatch in its center. These probe pairs, called the Perfect Match probe (PM) and the Mismatch probe (MM), allow the quantitation and subtraction of signals caused by non-specific cross-hybridization. The difference in hybridization signals between the partners, as well as their intensity ratios, serve as indicators of specific target abundance (website at http://www.affymetrix.com/technology/design/index.affx). In addition to allowing hybridization to the most specific regions of nucleotides, the use of short oligonucleotides also allows the representation of multiple regions of a single gene in multiple spots, thus reducing the chance of false positives (22). The main advantage of the GeneChip is its ability to measure the absolute expression of genes in cells or tissues (17). GeneChip technology also eliminates handling of bacterial libraries, amplification of sequences and the risk of cross contamination (22). Oligonucleotide microarrays like the GeneChip allow for the differential detection of gene family members or alternative transcripts that can not be distinguished using cDNA microarrays (2). The entire human genome can now be analyzed on a single array. The GeneChip Human Genome U133 Plus 2.0 array contains 1.3 million distinct oligonucleotides and was used to analyze the expression levels of over 47,000 transcripts as well as variants, including over 30,000 well-characterized human genes (23). In the sample preparation, one or two amplification rounds are used to generate cRNA after reverse transcription. This procedure can be carried out with significantly less starting material than is required for other methods, and the protocols use as little as 50 ng of total RNA, allowing analysis of samples that are small or otherwise difficult to obtain (22). Biotinylated nucleotides are used and the hybridized microarrays are stained with fluorescence-labeled streptavidin (22).
Other commercially available oligonucleotide microarrays are prepared using inkjet technology (Rossetta/Agilent) (24). This system was first described as a flexible system for gene expression profiling using arrays of tens of thousands of oligonucleotides synthesized in situ which are ink-jet printed using standard phosphoramidite chemistry methods. Hughes et al. reported that 60-mer oligonucleotides reliably detect transcript ratios at one copy per cell in complex biological samples, and that ink-jet arrays are compatible with several sample amplification and labeling techniques (24). In the sample preparation, different fluorescent-labeled nucleotides (Cy3/Cy5) can be incorporated into the cRNAs in Rosetta arrays, similar to cDNA microarrays.
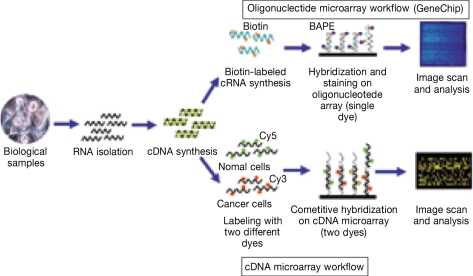
cDNA microarrays are made by spotting cDNAs, usually PCR-amplified sequences from bacterial libraries, onto glass slides (25). cDNA microarrays comprise relatively long DNA molecules immobilized on a solid surface and are mostly used for large-scale screening and expression studies. cDNA microarrays can not be used for mutation or genotyping analysis, which should be performed using oligonucleotide microarrays (2). Spot sizes range from 80~150 µm in diameter, and arrays can contain up to 80,000 spots (17). In terms of sample preparation, RNA from cells is reverse transcribed into cDNA, which is then fluorescently or radioactively labeled and used to probe a predetermined DNA set. Two different fluorescent dyes (usually Cy5 and Cy3) are used for cDNA microarray analysis, and a typical analysis may consist of a normal tissue sample being labeled with a green dye and a cancer tissue sample being labeled with a red dye. If both samples bind to the same target on a chip, a yellow signal is obtained, and a scanner is used to assess differing red/green/yellow emissions (26). The greater the hybridization signal from probe-DNA binding, the higher the concentration of the RNA within the original sample (27). In the example of target preparations, total RNA extracted from cells is fluorescently labeled by oligo dT-primed reverse transcription using nucleotides tagged with either Cy3 or Cy5. The unincorporated fluor-dUTPs are removed, the Cy3 and Cy5 probes combined, and then mixed with blockers. Agarose gel electrophoresis is usually used to assess the quality and quantity of extracted RNA. However, when the amount of RNA is limited, RNA quality and quantity can be assessed using the microcapillary-based Bionalyzer (Agilent Technologies), which can analyze as little as 5 ng of total RNA (17). The target mixture is hybridized to the probes on the microarrays for 16~24 hours, the array is then washed and scanned (20). Most microarray amplification methods make use of a linear-based amplification method using T7 RNA polymerase, resulting in amplified RNA (aRNA). Microarray studies that use the aRNA synthesis protocol require one amplification round, allowing as little as 10 ng or 1,000 cells to be used for the initial input (27,28) (Fig. 2).
The RET oligonucleotide microarray for MEN2A, MEN2B and FMTC syndromes was first developed by our group and we provide a free genetic screening of the RET gene (8). β-catenin and K-ras oligonucleotide microarrays were also developed for mutation screening of various cancer types (14,29). High throughput SNP analysis is also available using GeneChip microarrays (30,31). BRCA1 and BRCA2 (32,33), ATM (5) and p53 (34) oligonucleotide microarrays were developed for use in mutation analysis in cancer research (Fig. 3).
The gene expression profile of a cell determines its function, phenotype and response to stimuli. Thus, gene expression profiles can help elucidate cellular functions, biochemical pathways and regulatory mechanisms (2). Gene expression analysis using either oligonucleotide or cDNA microarrays is usually for "class comparison", "class prediction" and "class discovery" (35~37). Although cancer classification has improved over the past 30 years, there has been no general approach for identifying new cancer classes (class discovery) or for assigning tumors to known classes (class prediction) (37).
This type of study examines whether expression profiles differ between classes and, if so, attempts to identify the differentially expressed genes (35). Thus, these studies aim to identify genes differentially expressed among predefined classes of samples. Hedenfalk et al. (38) hypothesized that the genes expressed by two types of tumors (BRCA1 mutation carriers vs. BRCA2 mutation carriers) are distinctive. An analysis of variance between the levels of gene expression and the genotypes of the samples identified 176 genes that were differentially expressed in tumors with BRCA1 mutations and tumors with BRCA2 mutations. Yuan et al. (39) employed cDNA microarrays to identify patterns of gene expression among colorectal cancer cell lines and to directly compare lines with and without microsatellite instability. Multiple differential expression patterns were identified.
Another application of gene expression analysis using high density microarrays is to predict the biological group, diagnostic category or prognostic stage of a patient based on an expression profile from diseased tissue (35). A good example of class prediction is a recent study by Lossos et al. (40). That study examined 36 genes whose expression had been reported to predict survival in diffuse large-B-cell lymphoma. The genes that were the strongest predictors were LMO2, BCL6, FN1, CCND2, SCYA3 and BCL2. They found that measuring the expression of these six genes was sufficient to predict overall survival in diffuse large-B-cell lymphoma patients (40). A gene-expression signature was also used as a predictor of survival in breast cancer (41). In that study, microarray analysis was used to determine a previously-established 70-gene prognosis profile in a series of 295 consecutive patients with primary breast carcinomas. Patients were then classified as having a gene-expression signature associated with either a poor prognosis or a good prognosis (41). A similar approach for predicting breast cancer outcomes using gene expression profiling was reported by Huang et al. (42). Kihara et al. (43) demonstrated the feasibility of gene expression profiling using microarrays to predict survival in esophageal cancer patients receiving 5-FU-based adjuvant chemotherapy (43). Gene expression profiling-based prediction of response of colon carcinoma cells to 5-fluorouracil and camptothecin was also reported (44). Recently, Kim et al. (45) undertook a prospective study to identify correlations between gene expression and clinical resistance to 5-FU/cisplatin. Using an Affymetrix U133A microarray, those authors compared expression profiles from gastric cancer endoscopic biopsy specimens obtained during a chemosensitive state (partial remission after 5-FU/ cisplatin) with those obtained during a refractory state (disease progression). Using 119 discriminating probes and a cross-validation approach, they were able to correctly identify the chemo-responsiveness of 7 pairs of training samples and 1 independent test pair.
Class discovery in cancer research determines whether discrete subsets of a disease entity can be defined based on the gene expression profiles (35). Cluster analysis is suitable for class discovery, but is generally not appropriate for class comparison or class prediction (35). It is often used to identify clues regarding the heterogeneity of disease pathogenesis using class discovery analysis (36). Using a DNA microarray, Alizadeh et al. determined that diffuse large B-cell lymphomas (DLBCL), the most common subtype of non-Hodgkin's lymphoma, are clinically heterogeneous (46). They identified two molecularly distinct forms of DLBCL which had gene expression patterns indicative of different stages of B-cell differentiation. One type expressed genes characteristic of germinal centre B cells, while the second type expressed genes normally induced during in vitro activation of peripheral blood B cells. Patients with germinal centre B-like DLBCL had a significantly better overall survival than those with activated B-like DLBCL (46). Bittner et al. (47) reported global transcript analysis could identify unrecognized subtypes of cutaneous melanoma and could predict experimentally verifiable phenotypic characteristics that might be of importance in disease progression.
The above three classifications of class comparison, class prediction and class discovery are based on the criteria of Simon et al. (35,36) and Golub et al. (37). However, it is difficult to classify all gene expression analysis studies using microarrays into these three groups. Thus, I introduced a classification called 'Identification of biomarkers'. As many microarray analyses are aimed at finding significant biomarkers, many studies can be placed in this category. Some studies classified as belonging to one of the above three classifications can also belong to this category. Our group published gene expression profiling associated with multi-drug (5-FU, doxorubicin and cisplatin) resistance in human gastric cancer cells using Affymetrix U133A microarrays (48). A major obstacle in chemotherapy is treatment failure due to anticancer drug resistance. The gene expression patterns of 10 chemoresistant gastric cancer cell lines were compared with those of four parent cell lines. We identified over 250 genes differentially expressed in 5-fluorouracil-, cisplatin-, or doxorubicin-resistant gastric cancer cell lines. Our expression analysis also identified eight multidrug resistance candidate genes that were associated with resistance to two or more of the tested chemotherapeutic agents. Among these, midkine (MDK), a heparin-binding growth factor, was overexpressed in all drug-resistant cell lines, strongly suggesting that MDK might contribute to multidrug resistance in gastric cancer cells. Chun et al. performed a similar study to identify markers in drug-resistance. To gain insight into clinically relevant mechanisms of irinotecan resistance, those authors performed oligonucleotide microarray analysis on paired malignant effusion samples obtained from eight gastric cancer patients treated with irinotecan. When differences in the expression of genes were examined, five isoforms of the metallothionein family were identified as having higher signal log ratios in five non-responders compared to their ratios in three responders. These findings collectively suggest that irinotecan-induced up-regulation of metallothionein might be associated with irinotecan resistance in patients with gastric cancer, although it remains to be confirmed in a larger data set. Wreesmann et al. (50) identified MUC1 as an independent prognostic marker of papillary thyroid cancer. Koopmann et al. (51) reported that serum macrophage inhibitory cytokine 1 was a marker of pancreatic and other periampullary cancers using oligonucleotide microarrays analysis, in situ hybridization and immunohistochemistry.
Microarray technologies are becoming more important in cancer research as cancers result from the accumulation of many genetic and epigenetic changes. Microarrays are being increasingly used for diagnostic classification of cancers. Comprehensive and high throughput genetic analysis is an inevitable research tool in cancer research. However, there are some drawbacks regarding routine use of microarrays. The cost of microarray experiments is high, and experimental steps need to be more robust. Standardization of microarrays and experimental protocols is also important for comparing data between research groups. Data analysis tools and methods also need to be developed. Despite these issues it is clear that microarray technology will be a basic tool in future cancer research. Early cancer diagnosis will be performed using oligonucleotide microarrays, and prognosis after chemo- or radiotherapy may be predicted by gene expression profiling. With conventional histopathological data, gene expression analysis using microarrays will help researchers find significant answers for questions surrounding cancer. Progressive bioinformatics tools will further refine the power of microarray technologies.
References
1. Schena M, Davis RW. Schena M, editor. Technology standards for microarray research. Microarray biochip technology. 2000. Massachusetts: Eaton publishing;p. 1–18.
2. Russo G, Zegar C, Giordano A. Advantages and limitations of microarray technology in human cancer. Oncogene. 2003; 22:6497–6507. PMID: 14528274.

3. Yershov G, Barsky V, Belgovskiy A, Kirillov E, Kreindlin E, Ivanov I, Parinov S, Guschin D, Drobishev A, Dubiley S, Mirzabekov A. DNA analysis and diagnostics on oligonucleotide microchips. Proc Natl Acad Sci USA. 1996; 93:4913–4918. PMID: 8643503.

4. Wikman FP, Lu ML, Thykjaer T, Olesen SH, Andersen LD, Cordon-Cardo C, Orntoft TF. Evaluation of the performance of a p53 sequencing microarray chip using 140 previously sequenced bladder tumor samples. Clin Chem. 2000; 46:1555–1561. PMID: 11017932.

5. Hacia JG, Sun B, Hunt N, Edgemon K, Mosbrook D, Robbins C, Fodor SP, Tagle DA, Collins FS. Strategies for mutational analysis of the large multiexon ATM gene using high-density oligonucleotide arrays. Genome Res. 1998; 8:1245–1258. PMID: 9872980.

6. Kim IJ, Kang HC, Park JH, Shin Y, Ku JL, Yoo BC, Park JG. Hardiman G, editor. Development and application of an oligonucleotide microarray for mutational analysis. Microarrays methods and applications. 2003. Pennsylvania: DNA press;p. 250–271.
7. Wen WH, Bernstein L, Lescallett J, Beazer-Barclay Y, Sullivan-Halley J, White M, Press MF. Comparison of TP53 mutations identified by oligonucleotide microarray and conventional DNA sequence analysis. Cancer Res. 2000; 60:2716–2722. PMID: 10825146.
8. Kim IJ, Kang HC, Park JH, Ku JL, Lee JS, Kwon HJ, Yoon KA, Heo SC, Yang HY, Cho BY, Kim SY, Oh SK, Youn YK, Park DJ, Lee MS, Lee KW, Park JG. RET oligonucleotide microarray for the detection of RET mutations in multiple endocrine neoplasia type 2 syndromes. Clin Cancer Res. 2002; 8:457–463. PMID: 11839664.
9. Lopez-Crapez E, Livache T, Marchand J, Grenier J. K-ras mutation detection by hybridization to a polypyrrole DNA chip. Clin Chem. 2001; 47:186–194. PMID: 11159765.

10. Gitan RS, Shi H, Chen CM, Yan PS, Huang TH. Methylation-specificoligonucleotide microarray: a new potential for high-throughput methylation analysis. Genome Res. 2002; 12:158–164. PMID: 11779841.
11. Abraham SC, Nobukawa B, Giardiello FM, Hamilton SR, Wu TT. Sporadic fundic gland polyps: common gastric polyps arising through activating mutations in the beta-catenin gene. Am J Pathol. 2001; 158:1005–1010. PMID: 11238048.
12. Abraham SC, Montgomery EA, Giardiello FM, Wu TT. Frequent beta-catenin mutations in juvenile nasopharyngeal angiofibromas. Am J Pathol. 2001; 158:1073–1078. PMID: 11238055.
13. Saegusa M, Okayasu I. Frequent nuclear beta-catenin accumulation and associated mutations in endometrioid-type endometrial and ovarian carcinomas with squamous differentiation. J Pathol. 2001; 194:59–67. PMID: 11329142.
14. Kim IJ, Kang HC, Park JH, Shin Y, Ku JL, Lim SB, Park SY, Jung SY, Kim HK, Park JG. Development and applications of a beta-catenin oligonucleotide microarray: beta-catenin mutations are dominantly found in the proximal colon cancers with microsatellite instability. Clin Cancer Res. 2003; 9:2920–2925. PMID: 12912937.
15. Samowitz WS, Curtin K, Schaffer D, Robertson M, Leppert M, Slattery ML. Relationship of Ki-ras mutations in colon cancers to tumor location, stage, and survival: a population-based study. Cancer Epidemiol Biomarkers Prev. 2000; 9:1193–1197. PMID: 11097226.
16. Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, et al. Kirsten ras mutations in patients with colorectal cancer: the 'RASCAL II' study. Br J Cancer. 2001; 85:692–696. PMID: 11531254.
17. Macgregor PF, Squire JA. Application of microarrays to the analysis of gene expression in cancer. Clin Chem. 2002; 48:1170–1177. PMID: 12142369.

18. Warrington JA, Dee S, Trulson M. Schena M, editor. Large-scale genomic analysis using affymetrix GeneChip probe arrays. Microarray Biochip Technology. 2000. Massachusetts: Eaton Publishing;p. 201–220.
19. Chou WH, Yan FX, Robbins-Weilert DK, Ryder TB, Liu WW, Perbost C, Fairchild M, de Leon J, Koch WH, Wedlund PJ. Comparison of two CYP2D6 genotyping methods and assessment of genotype-phenotype relationships. Clin Chem. 2003; 49:542–551. PMID: 12651805.

20. Khan J, Saal LH, Bittner ML, Chen Y, Trent JM, Meltzer PS. Expression profiling in cancer using cDNA microarrays. Electrophoresis. 1999; 20:223–229. PMID: 10197427.

21. Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, Brown EL. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996; 14:1675–1680. PMID: 9634850.

22. Schmidt U, Begley CG. Cancer diagnosis and microarrays. Int J Biochem Cell Biol. 2003; 35:119–124. PMID: 12479861.

23. Jain KK. Applications of biochips: from diagnostics to personalized medicine. Curr Opin Drug Discov Devel. 2004; 7:285–289.
24. Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, Lefkowitz SM, Ziman M, Schelter JM, Meyer MR, Kobayashi S, Davis C, Dai H, He YD, Stephaniants SB, Cavet G, Walker WL, West A, Coffey E, Shoemaker DD, Stoughton R, Blanchard AP, Friend SH, Linsley PS. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol. 2001; 19:342–347. PMID: 11283592.

25. Schulze A, Downward J. Navigating gene expression using microarrays--a technology review. Nat Cell Biol. 2001; 3:E190–E195. PMID: 11483980.
26. Kurian KM, Watson CJ, Wyllie AH. DNA chip technology. J Pathol. 1999; 187:267–271. PMID: 10398077.
27. Glanzer JG, Haydon PG, Eberwine JH. Expression profile analysis of neurodegenerative disease: advances in specificity and resolution. Neurochem Res. 2004; 29:1161–1168. PMID: 15176473.

28. Sotiriou C, Powles TJ, Dowsett M, Jazaeri AA, Feldman AL, Assersohn L, Gadisetti C, Libutti SK, Liu ET. Gene expression profiles derived from fine needle aspiration correlate with response to systemic chemotherapy in breast cancer. Breast Cancer Res. 2002; 4:R3. PMID: 12052255.

29. Park JH, Kim IJ, Kang HC, Shin Y, Park HW, Jang SG, Ku JL, Lim SB, Jeong SY, Park JG. Oligonucleotide microarray-based mutation detection of the K-ras gene in colorectal cancers using Competitive DNA Hybridization (CDH). Clin Chem. 2004; 50:1688–1691. PMID: 15331511.
30. Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, Kruglyak L, Stein L, Hsie L, Topaloglou T, Hubbell E, Robinson E, Mittmann M, Morris MS, Shen N, Kilburn D, Rioux J, Nusbaum C, Rozen S, Hudson TJ, Lander ES, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998; 280:1077–1082. PMID: 9582121.

31. Mei R, Galipeau PC, Prass C, Berno A, Ghandour G, Patil N, Wolff RK, Chee MS, Reid BJ, Lockhart DJ. Genome-wide detection of allelic imbalance using human SNPs and high-density DNA arrays. Genome Res. 2000; 10:1126–1137. PMID: 10958631.

32. Hacia JG, Brody LC, Chee MS, Fodor SP, Collins FS. Detection of heterozygous mutations in BRCA1 using high density oligonucleotide arrays and two-colour fluorescence analysis. Nat Genet. 1996; 14:441–447. PMID: 8944024.

33. Favis R, Day JP, Gerry NP, Phelan C, Narod S, Barany F. Universal DNA array detection of small insertions and deletions in BRCA1 and BRCA2. Nat Biotechnol. 2000; 18:561–564. PMID: 10802632.
34. Ahrendt SA, Halachmi S, Chow JT, Wu L, Halachmi N, Yang SC, Wehage S, Jen J, Sidransky D. Rapid p53 sequence analysis in primary lung cancer using an oligonucleotide probe array. Proc Natl Acad Sci USA. 1999; 96:7382–7387. PMID: 10377423.

35. Simon R. Diagnostic and prognostic prediction using gene expression profiles in high-dimensional microarray data. Br J Cancer. 2003; 89:1599–1604. PMID: 14583755.

36. Simon R, Radmacher MD, Dobbin K, McShane LM. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J Natl Cancer Inst. 2003; 95:14–18. PMID: 12509396.

37. Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999; 286:531–537. PMID: 10521349.

38. Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi OP, Wilfond B, Borg A, Trent J. Gene-expression profiles in hereditary breast cancer. N Engl J Med. 2001; 344:539–548. PMID: 11207349.

39. Yuan Z, Sotsky Kent T, Weber TK. Differential expression of DOC-1 in microsatellite-unstable human colorectal cancer. Oncogene. 2003; 22:6304–6310. PMID: 13679870.

40. Lossos IS, Czerwinski DK, Alizadeh AA, Wechser MA, Tibshirani R, Botstein D, Levy R. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med. 2004; 350:1828–1837. PMID: 15115829.

41. Bvan de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictorof survival in breast cancer. N Engl J Med. 2002; 347:1999–2009. PMID: 12490681.
42. Huang E, Cheng SH, Dressman H, Pittman J, Tsou MH, Horng CF, Bild A, Iversen ES, Liao M, Chen CM, West M, Nevins JR, Huang AT. Gene expression predictors of breast cancer outcomes. Lancet. 2003; 361:1590–1596. PMID: 12747878.

43. Kihara C, Tsunoda T, Tanaka T, Yamana H, Furukawa Y, Ono K, Kitahara O, Zembutsu H, Yanagawa R, Hirata K, Takagi T, Nakamura Y. Prediction of sensitivity of esophageal tumors to adjuvant chemotherapy by cDNA microarray analysis of gene-expression profiles. Cancer Res. 2001; 61:6474–6479. PMID: 11522643.
44. Mariadason JM, Arango D, Shi Q, Wilson AJ, Corner GA, Nicholas C, Aranes MJ, Lesser M, Schwartz EL, Augenlicht LH. Gene expression profiling-based prediction of response of colon carcinoma cells to 5-fluorouracil and camptothecin. Cancer Res. 2003; 63:8791–8812. PMID: 14695196.
45. Kim HK, Choi IJ, Kim HS, Kim JH, Kim E, Park IS, Chun JH, Kim IH, Kim IJ, Kang HC, Park JH, Bae JM, Lee JS, Park JG. DNA microarray analysis of the correlation between gene expression patterns and acquired resistance to 5-FU/cisplatin in gastric cancer. Biochem Biophys Res Commun. 2004; 316:781–789. PMID: 15033468.

46. Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000; 403:503–511. PMID: 10676951.

47. Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, Sampas N, Dougherty E, Wang E, Marincola F, Gooden C, Lueders J, Glatfelter A, Pollock P, Carpten J, Gillanders E, Leja D, Dietrich K, Beaudry C, Berens M, Alberts D, Sondak V. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000; 406:536–540. PMID: 10952317.

48. Kang HC, Kim IJ, Park JH, Shin Y, Ku JL, Jung MS, Yoo BC, Kim HK, Park JG. Identification of genes with differential expression in acquired drug-resistant gastric cancer cells using high-density oligonucleotide microarrays. Clin Cancer Res. 2004; 10:272–284. PMID: 14734480.

49. Chun JH, Kim HK, Kim E, Kim IH, Kim JH, Chang HJ, Choi IJ, Lim HS, Kim IJ, Kang HC, Park JH, Bae JM, Park JG. Increased expression of metallothionein is associated with irinotecan resistance in gastric cancer. Cancer Res. 2004; 64:4703–4706. PMID: 15256434.

50. Wreesmann VB, Sieczka EM, Socci ND, Hezel M, Belbin TJ, Childs G, Patel SG, Patel KN, Tallini G, Prystowsky M, Shaha AR, Kraus D, Shah JP, Rao PH, Ghossein R, Singh B. Genome-wide profiling of papillary thyroid cancer identifies MUC1 as an independent prognostic marker. Cancer Res. 2004; 64:3780–3789. PMID: 15172984.

51. Koopmann J, Buckhaults P, Brown DA, Zahurak ML, Sato N, Fukushima N, Sokoll LJ, Chan DW, Yeo CJ, Hruban RH, Breit SN, Kinzler KW, Vogelstein B, Goggins M. Serum macrophage inhibitory cytokine 1 as a marker of pancreatic and other periampullary cancers. Clin Cancer Res. 2004; 10:2386–2392. PMID: 15073115.

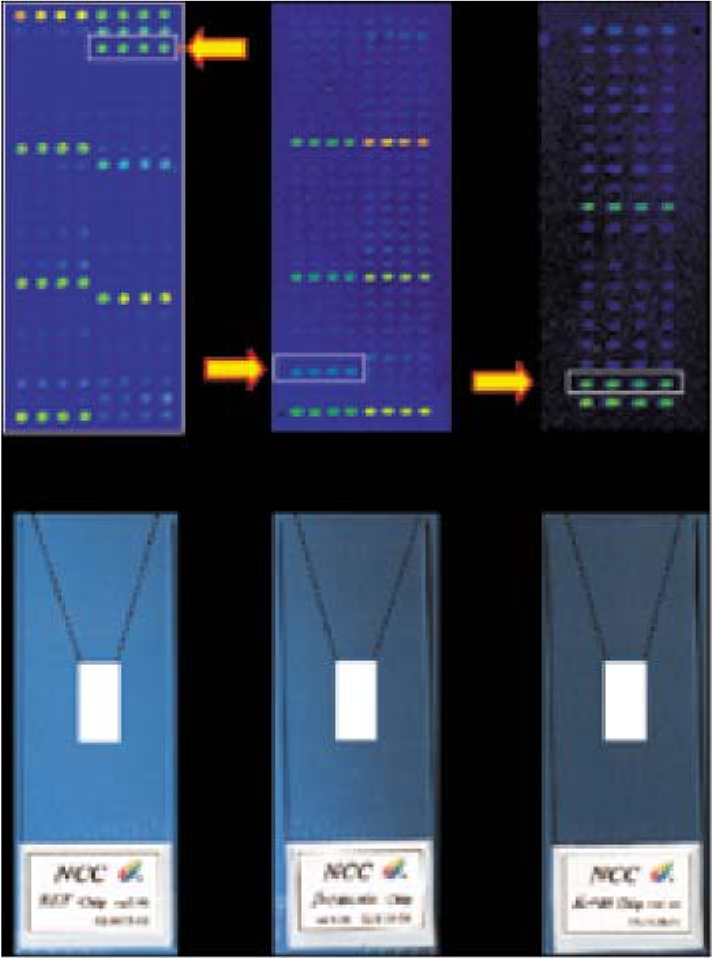
Fig. 1
Oligonucleotide microarrays for mutation analysis. Three microarrays were developed by the National Cancer Center in order to detect mutations in the RET, β-catenin and K-ras genes. The arrows indicate different types of mutations in DNA extracted from cancer tissues and blood samples.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download