Abstract
Background:
Isocitrate dehydrogenase (IDH) catalyzes the oxidative decarboxylation of isocitrate to yield α-ketoglutarate (α-KG) with production of reduced nicotinamide adenine dinucleotide (NADH). Dysfunctional IDH leads to reduced production of α-KG and NADH and increased production of 2-hydroxyglutarate, an oncometabolite. This results in increased oxidative damage and stabilization of hypoxia-inducible factor α, causing cells to be prone to tumorigenesis.
Methods:
This study investigated IDH mutations in 61 Ewing sarcoma family tumors (ESFTs), using a pentose nucleic acid clamping method and direct sequencing.
Results:
We identified four cases of ESFTs harboring IDH mutations. The number of IDH1 and IDH2 mutations was equal and the subtype of IDH mutations was variable. Clinicopathologic analysis according to IDH mutation status did not reveal significant results.
Conclusions:
This study is the first to report IDH mutations in ESFTs. The results indicate that ESFTs can harbor IDH mutations in previously known hot-spot regions, although their incidence is rare. Further validation with a larger case-based study would establish more reliable and significant data on prevalence rate and the biological significance of IDH mutations in ESFTs.
Ewing sarcoma (ES) is the second most common primary bone sarcoma of those that typically develop in children and young adults. It is also called ES family tumor (ESFT) and includes extraskeletal ES and primitive neuroectodermal tumor [1]. ESFT is an aggressive tumor with metastases present at diagnosis in 20%–25% of cases. With current therapeutic options, the 5-year survival rate for non-metastatic disease is as high as 70%. However, survival for patients who have metastasis is approximately 20%, and for those who develop relapsed or refractory disease, the survival rate is less than 10%. There is a need to identify alternative therapeutic agents that appropriately target the biomolecular mechanisms of this disease [1].
There is a group of tumors and tumor syndromes that carry mutations in metabolic enzymes involved in the tricarboxylic acid cycle, especially enzymes in the isocitrate dehydrogenase (IDH) family. IDH catalyzes the oxidative decarboxylation of isocitrate to yield α-ketoglutarate (α-KG) with production of reduced nicotinamide adenine dinucleotide (NADH). Dysfunctional IDH leads to reduced production of α-KG and NADH and increased production of 2-hydroxyglutarate, an oncometabolite. Together, this results in increased oxidative damage and stabilization of hypoxia-inducible factor α, causing cells to be prone to tumorigenesis [2]. A functional study proved that IDH2 mutations in mesenchymal cells can induce malignant transformation [3]. Mutations in IDH1 are reported to cluster at a single hotspot locus (R132), whereas IDH2 mutations occur primarily at two loci (R140 and R172) [2]. Recurrent somatic IDH1/2 mutations have been described in gliomas and secondary glioblastomas [4]. Similar IDH1/2 mutations have been detected in acute myeloid leukemia [5] and myelodysplastic disorders [6]. Recently, IDH mutations have been reported in a large proportion of cartilaginous tumors [7], a small number of osteosarcomas [8], and in giant cell tumors [9]. The fact that these mutations appear to be present in these relatively common bone tumors led us to investigate IDH1/2 mutations in ESFTs.
Formalin-fixed, paraffin-embedded tissue samples from 61 patients with primary localized ESFTs were obtained in Korea, Brazil, and Argentina. Fifty-five of the 61 tissue samples were obtained by surgical biopsy and the other six were obtained by surgical excision. At the time of tissue sampling, none of the patients had a history of chemotherapy or radiation therapy and there was no evidence of metastatic disease. The disease was diagnosed according to World Health Organization (WHO) criteria [10]. Briefly, they are small round cell sarcomas showing diffuse membranous CD99 immunostaining, cytoplasmic periodic acid–Schiff staining, and EWSR1 gene translocation as demonstrated with fluorescence in situ hybridization (ZytoLight SPEC EWSR1 Dual Color Break Apart Probes, ZytoVision, Bremerhaven, Germany). However, lack of an EWSR1 gene translocation was not considered as grounds for exclusion if a tumor showed the typical immunophenotypes, which are inconsistent with other small round cell tumors in the differential diagnosis of diseases such as lymphoma and rhabdomyosarcoma.
After histological diagnosis, the patients received standard multidrug chemotherapy in combination with surgery. Data, including the follow-up period and overall survival, were available for 48 patients. During follow-up, assessment of distant metastasis was available in 38 patients. The patients were grouped into dead of disease, alive with disease, and no evidence of disease (NED). The classification of patients as NED was established when the follow-up period had passed more than 24 months. Our study protocol was reviewed and approved by the Kyung Hee University Institutional Review Board.
IDH1/2 mutations were tested using the pentose nucleic acid (PNA) Clamp IDH1/2 Mutation Detection Kit (Panagene, Daejeon, Korea). All reactions had a total reaction volume of 20 μL and contained template DNA, primer and PNA probe sets, and fluorescent polymerase chain reaction (PCR) master mix. All required reagents were included with the kit. Real-time PCR reactions of PNA-mediated clamping PCR were performed using a CFX 96 (Bio-Rad, Hercules, CA, USA). PCR cycling conditions were as follows: 5 minutes at 94ºC followed by 40 cycles of 94ºC for 30 seconds, 70ºC for 20 seconds, 63ºC for 30 seconds, and 72ºC for 30 seconds. In this assay, PNA probes and DNA primers were used together in the clamping reaction. Positive signals were detected by intercalation of fluorescent dye. The PNA probe, which is complementary to the wild-type sequence, suppresses amplification of the wild-type target. This suppression results in preferential amplification of mutant sequences by competitively inhibiting the binding of DNA primers to wild-type DNA. PCR efficiency was determined by measuring the threshold cycle (Ct) value. Ct values for control and mutant assays were obtained from fluorescent amplification plots. Calculations of the delta Ct (ΔCt) value were done as follows: ΔCt1=(Standard Ct)–(Sample Ct), ΔCt2=(Sample Ct)–(Non-PNA mix Ct). The gene was considered to be mutated when ΔCt1 values were more than 2.0. When ΔCt1 values were between 0 and 2, a ΔCt2 value was then calculated. The gene was considered to be mutated if the calculated ΔCt2 value was ≤4.
Genomic PCR for sequencing was performed in 20-μL volumes using 30 ng of template DNA and 2× Taq PCR Smart Mix (Solgent, Daejeon, Korea). The PCR primers used for IDH1/2 amplification were as follows: IDH1 forward primer (5'-CGGTCTTCAGAGAAGCCATT-3') and IDH1 reverse primer (5'-GCAAAATCACATTATTGCCAAC-3'). IDH2 forward primer (5'-CCAATGGAACTATCCG-3') and IDH2 reverse primer (5'-CTCCACCCTGGCCTACCTG-3'). PCR cycling commenced with a 10 minutes hold at 95ºC, followed by 40 cycles of 95ºC for 30 seconds, 58ºC for 40 seconds, and 72ºC for 60 seconds, terminating with 72ºC for 5 minutes. Each amplified product was purified using a PCR clean-up kit (Macherey-Nagel, Duren, Germany) and sequenced in duplicate, in both the forward and reverse directions, using a BigDye Terminator Kit (Applied Biosystems, Carlsbad, CA, USA) on an ABI Prism 3100 station (Applied Biosystems), according to the manufacturer’s instructions. Sequences were compared with the GenBank-archived sequence of human IDH1/2.
The primary antibody that is specific for the IDH1 R132H point mutation (1:200, Histonova DIA-H09, Dianova, Hamburg, Germany) was used for the samples bearing IDH1 mutations, revealed by either direct PCR or PNA clamping. The Bond Polymer Intense Detection System (Vision Biosystems, Melbourne, Australia) was used according to the manufacturer's instructions with minor modifications. Nuclei were counterstained with hematoxylin. Paraffin-embedded tissues of brain astrocytomas were used as a positive control.
Statistical analyses were performed using SPSS Software (SPSS Inc., Chicago, IL, USA). Pearson’s chi-square test or Fisher exact test were performed to determine correlations between IDH mutation status and clinicopathological parameters. Statistical significance was defined as a p-value less than .05.
Using the PNA clamping method, IDH1/2 mutations were detected in three of the 61 patients (5%). Of these three samples, two were IDH1 mutants and one sample was an IDH2 mutant. By direct sequencing, IDH1/2 mutations were detected in two of the 61 patients (3%), of which, one sample was an IDH1 mutant and one sample was an IDH2 mutant. In total, four cases out of 61 (6%) harbored IDH1/2 mutations by at least one of the two methods employed, and the numbers of IDH1 and IDH2 mutants were equal (Table 1).
Table 2 summarizes the clinicopathologic characteristics of the four mutant cases. In one of four cases, the IDH1 mutation was found by both the PNA clamping method and direct sequencing (case No. 1) (Fig. 1A). In two of four cases, the IDH1/2 mutation was found only by the PNA clamping method (cases Nos. 2 and 3). In one of four cases, examination by the PNA clamping method showed equivocal results, but direct sequencing showed an IDH2 mutation (case No. 4) (Fig. 1C). The overall concordance rate of both methods was over 95% (58 of 61) and the discordance rate was less than 5% (3 of 61). In mutant cases, the concordance rate was 25% (1 of 4) and the discordance rate was 75% (3 of 4), although case No. 4 showed equivocal results by the PNA clamping method. Immunohistochemistry with antibody to DIA-H09 in the four cases bearing IDH1/2 mutations showed positive reactions only in case No. 1 (Fig. 1B).
In the four cases bearing IDH1/2 mutations, three patients were Korean and one patient was Brazilian, and the male/female ratio was 1:1. Three of the four patients were in their second decade and one patient was in the fifth decade. There was no evidence of distant metastasis in all patients and only one patient died during follow-up.
Statistical analyses showed that IDH1/2 mutant cases had stronger associations with Korean patients than with South American patients (p=.009). There was no significant association between IDH1/2 mutations and any of the other characteristics of tumors or patients (Table 3).
Research on IDH1/2 mutations in human tumors has been active in recent years and has revealed that various tumors of different origins bear IDH1/2 mutations [2]. Following increased interest in IDH1/2 mutations in soft tissue tumors, a rudimentary study on sarcoma cell lines demonstrated IDH mutations in fibrosarcoma [11]. Subsequently, a study on chondrogenic tumors demonstrated IDH1/2 mutations in 81 of 145 (56%) cases with an IDH1:IDH2 mutation ratio of 10.6:1. This study also included the evaluation of IDH1/2 mutations in 222 osteosarcomas, 79 chordomas, and 25 ESFTs, and no mutations were found [7]. Therefore, IDH mutations are considered to be found exclusively in chondrogenic tumors. Furthermore, previous studies support the value of examining IDH mutations for the purpose of differentiating chondrosarcoma from chondroblastic osteosarcoma [12] and chordoma [13]. However, a recent study in Japan showed that three of 12 osteosarcomas (25%) [8] and 16 of 20 giant cell tumors (80%) harbor IDH2 mutations [9], suggesting the possibility of IDH1/2 mutations in various soft tissue tumors in addition to chondrogenic tumors.
We demonstrated that four of 61 ESFTs (6%) possessed IDH mutations. The PNA clamping method is known to be sensitive, rapid, and simple to perform and can detect mutant alleles when present at levels 100-fold lower than those of wild-type alleles. In contrast, the minimum percentage of mutant DNA required for analysis by direct sequencing is more than 25% [14]. The two cases harboring IDH mutations, found only by PNA clamping, might have had less than 25% mutant DNA. It is worth noting that three osteosarcomas bearing IDH mutations were found in Japanese [8], and three ESFT bearing IDH mutations were found in Korean patients. However, evaluation of the same tumors from American patients revealed no mutations [7]. Although it is still early to remark on the background responsible for these findings, it is possible that Asian populations may be predisposed to IDH mutations in these tumors and therefore should be further evaluated.
Variation in the most prevalent mutation type according to tumor has been observed. IDH1 R132H represents the most common type in gliomas [4], and IDH2 R140Q is exclusively found in acute myeloid leukemia [5]. Whereas IDH1 R132C represents the most common type in cartilaginous tumors [13], IDH2 R172S is the dominant type in osteosarcomas [8] and giant cell tumors [9]. In our study, ESFTs demonstrated equal numbers of IDH1 and IDH2 mutations in which one case of R132H and one case of R172K were found. A previous study on cartilaginous tumors demonstrated that IDH mutations are frequent in acral-based tumors without any other association with other factors [7]. IDH mutations in osteosarcomas and giant cell tumors did not show any association with other clinical parameters [8,9]. Our study also did not find a significant association between IDH mutations and clinical parameters of ESFTs.
In conclusion, our study is the first report to demonstrate IDH mutations in ESFTs. It provides evidence that ESFTs can harbor IDH mutations in previously known hot-spot regions, although its incidence is rare. To provide generalized knowledge, our study is still lacking enough data about these mutations in ESFTs. Further validation with a larger case-based study would establish more reliable and significant data on prevalence rate and the biological significance of IDH1/2 mutation status in ESFTs.
REFERENCES
1. van Maldegem AM, Hogendoorn PC, Hassan AB. The clinical use of biomarkers as prognostic factors in Ewing sarcoma. Clin Sarcoma Res. 2012; 2:7.

2. Schaap FG, French PJ, Bovée JV. Mutations in the isocitrate dehydrogenase genes IDH1 and IDH2 in tumors. Adv Anat Pathol. 2013; 20:32–8.
3. Lu C, Venneti S, Akalin A, et al. Induction of sarcomas by mutant IDH2. Genes Dev. 2013; 27:1986–98.
4. Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009; 360:765–73.
5. Abbas S, Lugthart S, Kavelaars FG, et al. Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: prevalence and prognostic value. Blood. 2010; 116:2122–6.
6. Abdel-Wahab O, Manshouri T, Patel J, et al. Genetic analysis of transforming events that convert chronic myeloproliferative neoplasms to leukemias. Cancer Res. 2010; 70:447–52.

7. Amary MF, Bacsi K, Maggiani F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011; 224:334–43.
8. Liu X, Kato Y, Kaneko MK, et al. Isocitrate dehydrogenase 2 mutation is a frequent event in osteosarcoma detected by a multi-specific monoclonal antibody MsMab-1. Cancer Med. 2013; 2:803–14.

9. Kato Kaneko M, Liu X, Oki H, et al. Isocitrate dehydrogenase mutation is frequently observed in giant cell tumor of bone. Cancer Sci. 2014; 105:744–8.

10. Fletcher CD, Bridge JA, Hogendoorn PC, Mertens F. WHO classification of tumours of soft tissue and bone. 4th ed. Lyon: IARC Press;2013.
11. Rasheed S, Nelson-Rees WA, Toth EM, Arnstein P, Gardner MB. Characterization of a newly derived human sarcoma cell line (HT-1080). Cancer. 1974; 33:1027–33.

12. Kerr DA, Lopez HU, Deshpande V, et al. Molecular distinction of chondrosarcoma from chondroblastic osteosarcoma through IDH1/2 mutations. Am J Surg Pathol. 2013; 37:787–95.
13. Arai M, Nobusawa S, Ikota H, Takemura S, Nakazato Y. Frequent IDH1/2 mutations in intracranial chondrosarcoma: a possible diagnostic clue for its differentiation from chordoma. Brain Tumor Pathol. 2012; 29:201–6.
14. Lee HJ, Xu X, Kim H, et al. Comparison of direct sequencing, PNA clamping-real time polymerase chain reaction, and pyrosequencing methods for the detection of EGFR mutations in non-small cell lung carcinoma and the correlation with clinical responses to EGFR tyrosine kinase inhibitor treatment. Korean J Pathol. 2013; 47:52–60.
Fig. 1.
The case No. 1 sample shows IDH1 R132H mutation by direct sequencing (A) and positive immunoreactivity with antibody clone H09 (B). The case No. 4 sample shows IDH2 R172K mutation (C).
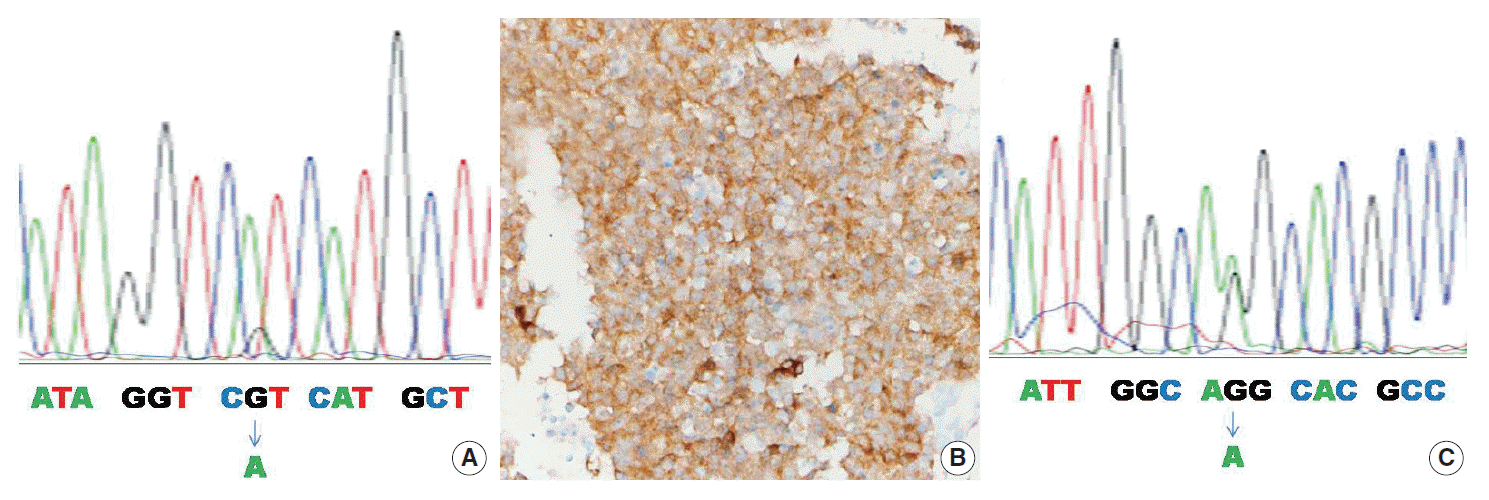
Table 1.
Results of IDH1/2 mutation analysis using PNA clamping and direct sequencing
| PNA clamping | Direct sequencing | |
|---|---|---|
| Wild type | 57 | 59 |
| Mutant | ||
| IDH1 | 2 | 1 |
| IDH2 | 1 | 1 |
| Equivocal | 1 | 0 |
Table 2.
Summary of four cases bearing IDH1/2 mutations, including clinicopathologic characteristics
Table 3.
Clinicopathologic analysis according to IDH1/2 mutation status




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download