Abstract
Background:
The conventional method for decalcification of bone specimens uses hydrochloric acid (HCl) and is notorious for damaging cellular RNA, DNA, and proteins, thus complicating molecular and immunohistochemical analyses. A method that can effectively decalcify while preserving genetic material is necessary.
Methods:
Pairs of bilateral bone marrow biopsies sampled from 53 patients were decalcified according to protocols of two comparison groups: EDTA versus HCl and RDO GOLD (RDO) versus HCl. Pairs of right and left bone marrow biopsy samples harvested from 28 cases were allocated into the EDTA versus HCl comparison group, and 25 cases to the RDO versus HCl comparison group. The decalcification protocols were compared with regards to histomorphology, immunohistochemistry, and molecular analysis. For molecular analysis, we randomly selected 5 cases from the EDTA versus HCl and RDO versus HCl groups.
Results:
The decalcification time for appropriate histomorphologic analysis was the longest in the EDTA method and the shortest in the RDO method. EDTA was superior to RDO or HCl in DNA yield and integrity, assessed via DNA extraction, polymerase chain reaction, and silver in situ hybridization using DNA probes. The EDTA method maintained intact nuclear protein staining on immunohistochemistry, while the HCl method produced poor quality images. Staining after the RDO method had equivocal results. RNA in situ hybridization using kappa and lambda RNA probes measured RNA integrity; the EDTA and RDO method had the best quality, followed by HCl.
Sampling bone tissue is usually performed for the diagnosis of hematologic malignancy, metastatic tumor, or primary bone tumor. The processing of bone specimens usually follows decalcification and microtome sectioning in pathology laboratories. Inorganic acids such as nitric acid or HCl are used in decalcification, and limit diagnostic options by damaging DNA and RNA. As a result, gene testing is usually not plausible with these decalcified bone specimens, even though certain cancers need further genetic studies for diagnostic and therapeutic purposes. Therefore, there is a growing need for new decalcification agents that adequately preserve DNA and RNA [1-3].
A variety of molecular testing techniques are necessary to diagnose hematologic malignancies. Fluorescence in situ hybridization, gene rearrangement studies of immunoglobulin and T cell receptor genes, and in situ hybridization for kappa and lambda light chains and Epstein-Barr virus–encoded small RNAs are commonly used molecular tools in diagnosis of hematologic malignancies. However, further molecular study from bone marrow biopsy specimens is often impossible due to DNA or RNA damage by decalcification. Furthermore, immunohistochemistry may be required for differentiating and subtyping hematolymphoid lesions in conjunction with histomorphologic features of paratrabecular, interstitial, intrasinusoidal, or intravascular aggregates within bone marrow structures. HCl degrades both protein quality and quantity, resulting in poor immunohistochemical staining that cannot be used for accurate diagnosis.
With consideration of these limitations, we evaluated modified bone marrow decalcification protocols and compared them to the HCl method.
This was a prospective study. To eliminate bias due to variables among cases, cases were enrolled when pairs of bilaterally biopsied bone marrow specimens were available. Among bone marrow specimens sampled from the right and left iliac crests between January 2013 and July 2014 at Gangnam Severance Hospital, 53 cases were finally included. For the 53 selected cases, 28 right and left bone marrow samples were allocated to the EDTA (Sigma-Aldrich, St. Louis, MO, USA) protocol and HCl (Calci-Clear Rapid, National Diagnostics, Atlanta, GA, USA) protocol, respectively. Samples from the 25 remaining cases were assigned to the RDO GOLD (RDO) group (Apex Engineering Products Corporation, Aurora, IL, USA) protocol and HCl protocol. Concentration, processing time, and temperature are summarized in Table 1.
To test DNA quality, five cases were randomly selected from each of the three groups (EDTA, HCl, and RDO). DNA was extracted, and the quantity and quality were confirmed using NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). The BRAF PNA clamping method (Panagene, Daejeon, Korea) and comparison with Ct values of internal controls were used to evaluate the efficacy of polymerase chain reaction (PCR) DNA amplification. To evaluate the efficacy of DNA in situ hybridization, HER2 dual color silver in situ hybridization (Ventana, Tucson, AZ, USA) was applied to 10 pairs of bone marrow samples. RNA in situ hybridization and immunohistochemical studies were performed prospectively. According to the potential differential diagnoses for suspicious lesions observed within bone marrow, appropriate RNA probes or protein antibodies were applied. For immunohistochemistry, the following primary antibodies were used and the details are summarized in Table 2: cyclin D1, Ki67, Bcl2, Bcl6, TdT, CD138, CD20, CD79a, CD3, CD5, CD23, CD10, CD30, and myeloperoxidase. To state the process, after deparaffinization and rehydration, the sections were incubated in BenchMark XT automated slide stainer (Ventana) for 16 minutes at 37°C and then counterstained with hematoxylin reagent. Two pathologists reviewed the hematoxylin and eosin (H&E), immunohistochemistry, and in situ hybridization slides. The quality of immunohistochemistry and RNA in situ hybridization were assessed using a 3-tiered grading scale: good, equivocal, or poor. HER2 silver in situ hybridization was assessed by detecting two signals of HER2 and CEP17 per nucleus from the normal hematopoietic cells of bone marrow.
The differences in variables were analyzed using the Mann-Whitney U test. All statistical analyses were carried out by SPSS ver. 20.0 for Windows (IBM Co., Armonk, NY, USA).
DNA quantity, purity, and Ct values of internal controls after real-time PCR in the EDTA versus HCl group and RDO versus HCl group are depicted in Table 3 and Fig. 1. Although differences were not statistically significant, the DNA yield of the EDTA protocol was about 2 times higher than the HCl protocol. In addition, the Ct value of the former protocol was significantly lower than that of the latter (p<.001) with the estimated difference being about 7. Furthermore, the Ct values of EDTA processed samples were lower than 30, demonstrating that the amount of intact DNA feasible for PCR with the EDTA protocol is better preserved by a factor of 2 [7] than the HCl protocol.
There were no significant differences between DNA yield and Ct values of RDO and HCl methods. The yield of extracted DNA after RDO decalcification was similar to that of HCl. The Ct values of both protocols were above 33, indicating that the amount of intact DNA feasible for PCR was very small.
The morphological comparison and quality assessment of H&E stain, HER2/CEP17 dual color silver in situ hybridization, kappa/lambda in situ hybridization, and immunohistochemistry studies were analyzed. The rates of high quality staining for each study were compared between the three protocols. The results are summarized in Table 4.
There was no difficulty in microtome dissection of 4 µm or less in thickness in any of the three methods. The morphological quality of H&E slide was similar in all three protocols, showing well-preserved histological features of the bone marrow (Fig. 2).
All five cases in the RDO versus HCl group had severe DNA breakdown on HER2/CEP17 dual color silver in situ hybridization, revealing no HER2 or CEP17 nuclear signal in bone marrow hematopoietic cells. However, in the EDTA versus HCl group, all 5 cases in the EDTA protocol showed two HER2 and CEP17 nuclear signals from almost all of the hematopoietic cells, whereas nearly no nuclear signal was detected in samples from the HCl protocol (Fig. 3).
Bone marrow specimens from the EDTA and RDO protocols that underwent kappa/lambda RNA in situ hybridization showed well preserved RNA signal in the nuclei of plasma cells, while those in the HCl group had severe breakdown of RNA signals (Fig. 4).
Nuclear proteins such as Ki67, cyclin D1, and TdT were relatively better preserved on immunohistochemistry with both EDTA and RDO protocols, while samples from the HCl protocol showed breakdown and lower quality of nuclear protein staining (Fig. 5A–H). Immunohistochemistry targeting the cytoplasmic membrane or cytoplasmic CD markers was well preserved in all three protocols (Fig. 5I–L).
There have been several studies comparing several types of decalcification protocols to date. Some retrospective studies compared the morphology of in situ hybridization or immunohistochemistry by using stored tissues containing bone. Other studies extracted nucleic acid from stored tissues containing bone and compared the quantity, purity, and Ct value of the DNA and/or RNA after real-time PCR [1-4]. Few studies have used specimens sampled with diagnostic purpose in clinical practice. We investigated the quality of nucleic acid and protein in decalcified bone marrow tissue employing several types of genetic tools, immunohistochemistry, and morphological assessments. In addition, we compared the effect of decalcification protocol while limiting bias variance, which may be caused by sampling from different patients and cell degeneration from long-term storage. We prospectively investigated pairs of bone marrow biopsy specimens from the same patients sampled for diagnostic purposes in a clinical setting. To our knowledge, this is one of the first studies utilizing clinical samples to assess decalcification protocols.
We compared the conventional protocol using HCl, the well-known alternative protocol using EDTA, and the new protocol using RDO.
All three methods had equally good performance with respect to microtome dissection and preservation of cytomorphologic and histomorphologic features. The EDTA protocol was superior in preserving nucleic acid (DNA and RNA), allowing for the feasibility of genetic studies, such as real time PCR and in situ hybridization. The EDTA protocol would be an appropriate option for genetic studies in bone-contained tissues. The present study also confirmed that the HCl protocols are inappropriate for genetic studies due to the severe damage of genetic material. The quality of nucleic acids in the RDO group was equivocal, but this method also didn’t seem to be suitable for genetic studies. In immunohistochemistry targeting nuclear protein, both EDTA and RDO were relatively superior to the HCl protocol. All three methods showed intact staining in immunohistochemistry targeting cytoplasmic membranes or cytoplasm. EDTA protocols seem to be the most appropriate in that comprehensive immunohistochemical stains can be applied to the specimens processed by this protocol. The feasibility of immunohistochemistry using specimens processed with the HCl protocol would be limited in many cases, especially when immunostaining for nuclear proteins is necessary. Immunohistochemical markers can be more widely applied in samples processed with the RDO protocol than with the HCl protocol, but diagnostic options are still more limited than the EDTA protocol.
In this study, the EDTA protocol was the most feasible method for several types of genetic studies and immunohistochemistry. Such advantages of EDTA decalcification protocol are already widely known [1,5-7]. The present study confirms again the superiority of the EDTA protocol in a wide range of ancillary tests for pathologic diagnosis. The potential of the RDO protocol is higher than the conventional HCl protocol; however, it is not superior to the EDTA protocol and thus, it cannot be considered an alternative to EDTA protocols, particularly when genetic studies are needed.
When choosing decalcification agents, cost-effectiveness and turn-around time from biopsy to final pathologic diagnosis are important issues. HCl costs less and requires less time than using EDTA. Small bone tissues like bone marrow take about three to twenty four hours for decalcification in EDTA protocol, but most of the other bone specimens need more time when using the same protocol. RDO protocols can shorten decalcification time, but this agent is also expensive.
In many cases, bone biopsies are needed for diagnosis of hematologic cancer, metastatic tumors, and primary bone sarcoma; these types of cancers usually need further gene-based diagnosis. Bone marrow biopsy is widely performed in patients with hematologic malignancies and pediatric sarcomas or blastomas. Therefore, preserving genetic materials is critical in handling bone marrow tissues, and cost and turn-around time may be less important in such cases. Preserving intact nucleic acid, as well as intact proteins, is critical to providing not only accurate pathologic diagnosis but also diverse therapeutic options to the patients. Considering this, in this study the EDTA protocol was the most appropriate method for handling bone marrow specimens. RDO may also be useful in that it requires less decalcification time and it enables more ancillary tests than HCl, but its usefulness is limited by less potential for genetic studies in processed samples than EDTA.
In this era of expanding genetic molecular study, better tissue handling methods are needed. The present study suggests an appropriate approach in handling bone marrow tissues, and this approach should be expanded to other types of tissue specimens.
ACKNOWLEDGMENTS
This study was supported by a faculty research grant of Yonsei University College of Medicine for 2014 (6-2014-0133).
REFERENCES
1. Singh VM, Salunga RC, Huang VJ, et al. Analysis of the effect of various decalcification agents on the quantity and quality of nucleic acid (DNA and RNA) recovered from bone biopsies. Ann Diagn Pathol. 2013; 17:322–6.

2. Reineke T, Jenni B, Abdou MT, et al. Ultrasonic decalcification offers new perspectives for rapid FISH, DNA, and RT-PCR analysis in bone marrow trephines. Am J Surg Pathol. 2006; 30:892–6.

3. Brown RS, Edwards J, Bartlett JW, Jones C, Dogan A. Routine acid decalcification of bone marrow samples can preserve DNA for FISH and CGH studies in metastatic prostate cancer. J Histochem Cytochem. 2002; 50:113–5.

4. Alers JC, Krijtenburg PJ, Vissers KJ, van Dekken H. Effect of bone decalcification procedures on DNA in situ hybridization and comparative genomic hybridization. EDTA is highly preferable to a routinely used acid decalcifier. J Histochem Cytochem. 1999; 47:703–10.

5. Adegboyega PA, Gokhale S. Effect of decalcification on the immunohistochemical expression of ABH blood group isoantigens. Appl Immunohistochem Mol Morphol. 2003; 11:194–7.

6. Castania VA, Silveira JW, Issy AC, et al. Advantages of a combined method of decalcification compared to EDTA. Microsc Res Tech. 2015; 78:111–8.

7. Wickham CL, Sarsfield P, Joyner MV, Jones DB, Ellard S, Wilkins B. Formic acid decalcification of bone marrow trephines degrades DNA: alternative use of EDTA allows the amplification and sequencing of relatively long PCR products. Mol Pathol. 2000; 53:336.
Fig. 1.
The quality, quantity, and feasibility of real time PCR study is compared between EDTA, RDO, and HCl protocols. The first row demonstrates EDTA versus HCl, and the second row RDO versus HCl (A, D, DNA yield; B, PCR result of EDTA; E, PCR result of RDO; C, F, PCR results of HCl). D, PCR result of RDO. PCR, polymerase chain reaction; EDTA, ethylenediaminetetraacetic acid disodium salt dehydrate; RDO, RDO GOLD; HCl, hydrochloric acid; PNA, peptide nucleic acid.
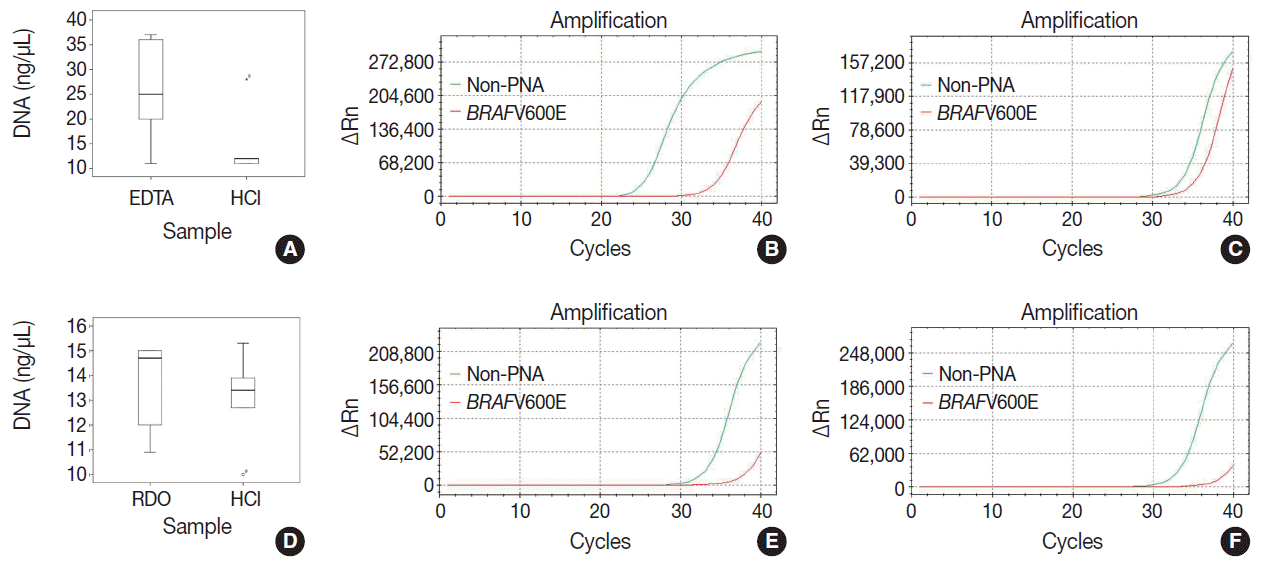
Fig. 2.
In EDTA versus HCl comparison of a pair of bone marrow sampled from the same patient (A–D, EDTA versus HCl group; A, B, EDTA protocol; C, D, HCl protocol), and in RDO versus HCl comparison of a pair of bone marrow sampled from the same patient (E–H, RDO versus HCl group; E, F, RDO protocol; G, H, HCl protocol), all the three methods of EDTA, RDO, and HCl protocols demonstrate intact and well-preserved histological features of bone marrow. EDTA, ethylenediaminetetraacetic acid disodium salt dehydrate; HCl, hydrochloric acid; RDO, RDO GOLD.
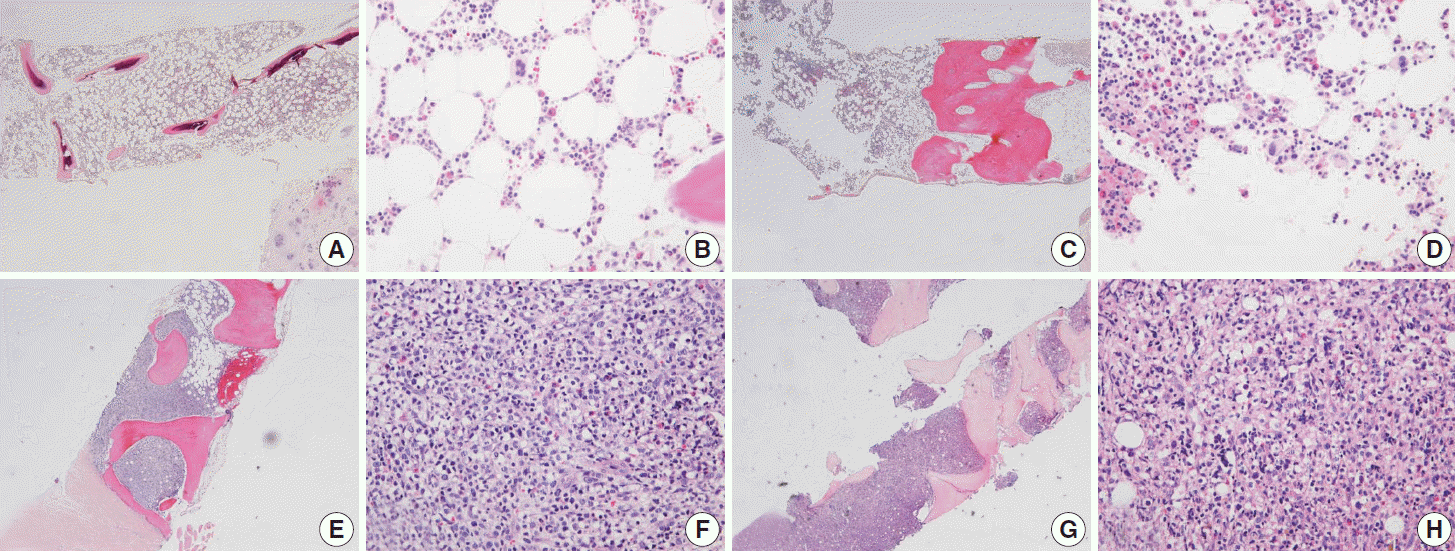
Fig. 3.
In HER2 dual color silver in situ hybridization study, almost every nucleus in cases of EDTA protocol demonstrates two intact signals of HER2 and CEP17, while cases of RDO or HCl protocol barely demonstrate HER2 or CEP17 signals from the nucleus due to the severe breakdown of DNA (A, B, comparison of EDTA versus HCl in a pair of bone marrow from the same patients; C, D, comparison of RDO versus HCl in a pair of bone marrow from the same patients). EDTA, ethylenediaminetetraacetic acid disodium salt dehydrate; RDO, RDO GOLD; HCl, hydrochloric acid.
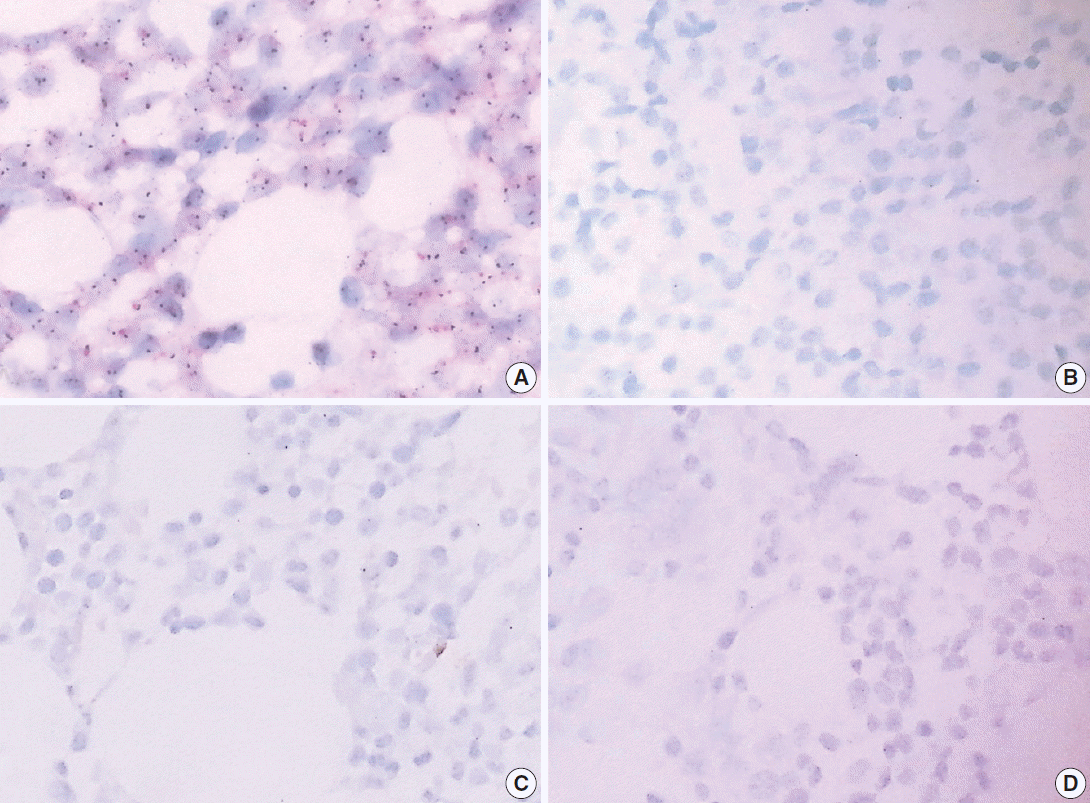
Fig. 4.
The quality of RNA is compared in kappa light chain (A–D) and lambda light chain (E–H) RNA in situ hybridization using a pair of bone marrow specimens from the same patient. In a case of kappa light chain-restricted plasma cell myeloma, EDTA protocol (A) reveals intact quality while HCl (B) protocol shows poor quality in kappa light chain RNA in situ hybridization. In a case of lambda light chain-restricted plasma cell myeloma, EDTA protocol (E) reveals intact quality while HCl (F) protocol shows poor quality. In a case of polyclonal plasma cell infiltration within the bone marrow, the EDTA and RDO protocol (C and G, respectively) reveal intact quality while HCl (D, H) protocol show poor quality in kappa and lambda light chain RNA in situ hybridization, respectively. EDTA, ethylenediaminetetraacetic acid disodium salt dehydrate; HCl, hydrochloric acid; RDO, RDO GOLD.
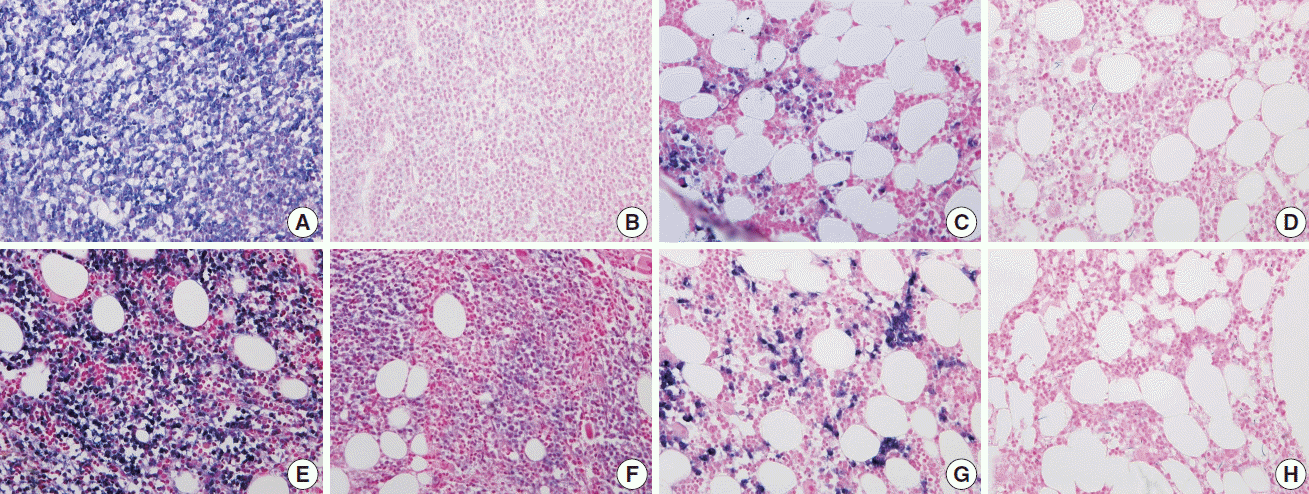
Fig. 5.
In immunohistochemistry of Ki67 (A–D), EDTA (A) shows intact quality while HCl (B) shows poor quality in a pair of bone marrow sample from the same patient. A similar result is noted in comparison of RDO (C) versus HCl (D). Nuclear staining of cyclin D1 is intact in EDTA (E), while it is not in HCl (F) of a paired bone marrow from the same patient. Nuclear staining of TdT is intact in RDO (G), while it is poor in HCl (H) of a paired bone marrow from the same patient. Cytoplasmic membrane staining of CD138 (I–L) reveals intact quality in all three protocols: EDTA (I), RDO (K), and HCl (J, L). EDTA, ethylenediaminetetraacetic acid disodium salt dehydrate; HCl, hydrochloric acid; RDO, RDO GOLD.
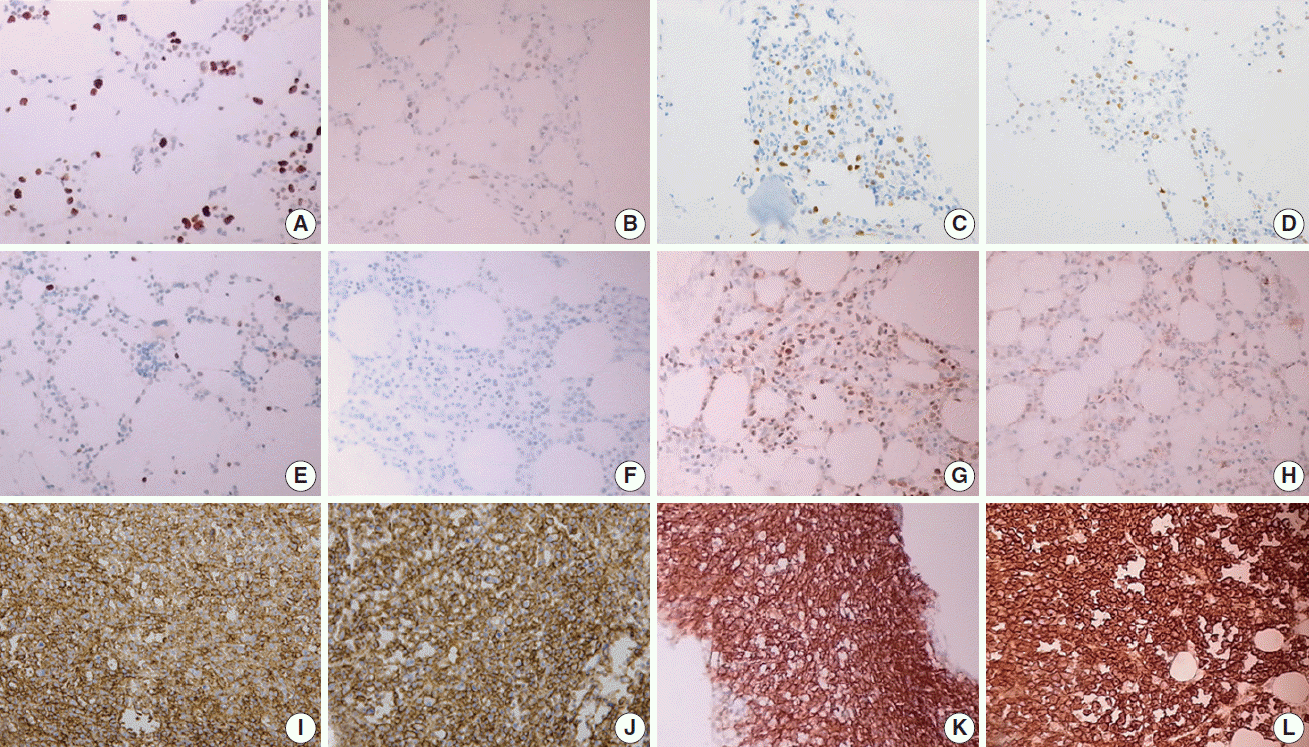
Table 1.
The decalcification protocols of three methods
| Solution | Processing time (hr) | Processing temperature |
|---|---|---|
| HCl (100%) | 3 | Room temperature |
| EDTA (12.5%) | 3 or 24a | Room temperature |
| RDO (100%) | 0.5–1 | Room temperature |
Table 2.
Primary antibodies used
| Product name | Dilution | Clonality | Clone | Company |
|---|---|---|---|---|
| Cyclin D1 (SP4) | 1:50 | Monoclonal | SP4 | LabVisiona |
| Ki67 | 1:1,000 | Monoclonal | MIB-1 | DAKOb |
| Bcl2 Bond-III | 1:50 | Monoclonal | bcl2/100/D5 | Novocastrac |
| Bcl6 | Prediluent | Monoclonal | LN22 | Novocastra |
| TdT | 1:100 | Polyclonal | - | Cell Marqued |
| CD138 | Prediluent | Monoclonal | ML15 | DAKO |
| CD20 | 1:400 | Monoclonal | L26 | Novocastra |
| CD79a (B cell) | 1:100 | Monoclonal | JCB117 | DAKO |
| CD3 | 1:200 | Monoclonal | SP7 | LabVision |
| CD5 | 1:100 | Monoclonal | 4C7 | Novocastra |
| CD23 | 1:100 | Monoclonal | SP23 | LabVision |
| CD10 | 1:75 | Monoclonal | 56C6 | Novocastra |
| CD30 | 1:50 | Monoclonal | Ber-H2 | DAKO |
| Myeloperoxidase | 1:2,000 | Polyclonal | - | DAKO |
Table 3.
Quantity and quality of DNA according to decalcification protocols
Table 4.
Comparison of the immunohistochemical results
| Item |
EDTA vs HCl |
RDO vs HCl |
||
|---|---|---|---|---|
| EDTA | HCl | RDO | HCl | |
| HER2/CEP17 SISH | 5/5 (100)a | 0/5 (0) | 0/5 (0) | 0/5 (0) |
| Kappa ISH | 7/7 (100) | 4/7 (57.1) | 2/3 (66.7) | 2/3 (66.7) |
| Lambda ISH | 5/5 (100) | 1/5 (20) | 2/2 (100) | 0/2 (0) |
| CyclinD1 | 9/9 (100) | 2/9 (22.2) | 3/3 (100) | 1/3 (33.3) |
| Ki67 | 9/9 (100) | 5/9 (55.6) | 10/10 (100) | 6/10 (45.5) |
| Bcl2 | 1/1 (100) | 1/1 (100) | 4/4 (100) | 4/4 (100) |
| Bcl6 | No data | No data | 1/1 (100) | 1/1 (100) |
| TdT | No data | No data | 1/1 (100) | 0/1 (0) |
| CD138 | 7/7 (100) | 7/7 (100) | 3/3 (100) | 3/3 (100) |
| CD20 | 9/9 (100) | 9/9 (100) | 9/9 (100) | 9/9 (100) |
| CD79a (B cell) | No data | No data | 2/2 (100) | 2/2 (100) |
| CD3 | 5/5 (100) | 5/5 (100) | 4/4 (100) | 4/4 (100) |
| CD5 | No data | No data | 3/3 (100) | 3/3 (100) |
| CD23 | No data | No data | 1/1 (100) | 1/1 (100) |
| CD10 | 1/1 (100) | 1/1 (100) | 1/1 (100) | 1/1 (100) |
| CD30 | 1/1 (100) | 1/1 (100) | No data | No data |
| Myeloperoxidase | 1/1 (100) | 1/1 (100) | 2/2 (100) | 2/2 (100) |
HCl, hydrochloric acid; EDTA, ethylenediaminetetraacetic acid disodium salt dehydrate; RDO, RDO GOLD; SISH, silver in situ hybridization; ISH, in situ hybridization.
a Case number showing intact stain result among overall case number stained with each item (%). Positive expression in the indicated tumor cells or internal controls was considered as intact stain. For example, positive expression of cyclin D1 in mantle cell lymphoma cells or endothelial cells was interpreted as intact stain result.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download