Abstract
Background
MicroRNA-552 (miR-552) has been reported to correlate with the development and progression of various cancers, including colorectal cancer (CRC). This study aimed to investigate miR-552 expression in cancer tissue samples compared to normal mucosal tissue and its role as a diagnostic or prognostic marker in CRC patients.
Methods
Normal mucosal tissues and primary cancer tissues from 80 surgically resected CRC specimens were used. Quantitative real-time polymerase chain reaction was performed for miR-552 and U6 small nuclear RNA to analyze miR-552 expression and its clinicopathological significance. Immunohistochemistry for p53 and phosphatase and tension homolog (PTEN) was performed to evaluate their association with miR-552 expression.
Results
miR-552 expression was significantly higher in primary cancer tissues compared to normal mucosal tissues (p<.001). The expression level of miR552 was inversely correlated with that of PTEN (p=.068) and p53 (p=.004). Survival analysis showed that high miR-552 expression was associated with worse prognosis but this was not statistically significant (p=.255). However, patients with CRC having high miR-552 expression and loss of PTEN expression had significantly worse prognosis than others (p=.029).
Colorectal cancer (CRC) is a disease with a high mortality rate affected by metastasis to other organs. The incidence of CRC has increased worldwide, making it the third most common cancer, next only to lung and breast cancers. Its mortality rate ranks second globally [1]. In South Korea, CRC is the second most common type of cancer after stomach cancer, caused by a westernized dietary pattern [2]. Moreover, its mortality rate has been rapidly increasing.
Previous studies on the mechanism of colorectal carcinogenesis have demonstrated the potential of microRNAs (miRNAs) as prognostic and diagnostic markers [3-5]. miRNAs are small non-coding, single-stranded RNA molecules consisting of 19–25 nucleotides capable of controlling hundreds of target genes [6,7]. A miRNA either disassembles the mRNA or hinders its translation to protein, preventing protein expression [8]. RNase III Dicer disassembles pre-miRNA into a mature double-stranded miRNA of approximately 22 nucleotides; thereafter, it combines with the RNA-indicated silencing complex, splitting the mRNA itself or preventing mRNA transcription [9]. miRNAs are also involved in the activity of over 50% of human genes by affecting gene expression and dysregulation, which prevents the formation and progression of tumors. Therefore, miRNAs are involved in formation or progression of various cancers [10]. For example, miRNA are major regulators of the Wnt/β-catenin signaling pathway during cell proliferation and survival [11-13]. Previous studies suggest that there are changes in miRNA expression in various cancer cells compared to normal cells [14-16]. Therefore, alterations in miRNA expression can be used for cancer diagnosis [17,18].
Previous studies have shown that miR-552 is involved in the carcinogenesis, progression, and metastasis of CRC [19-22]. It has been reported that miR-552 promotes the proliferation and invasion of CRC cells by directly targeting DACH1 through the Wnt/β-catenin signaling pathways [23]. miR-552 has been shown to activate the Wnt/β-catenin signaling pathway to regulate the expression of the tumor suppressors p53 and phosphatase and tension homolog (PTEN) [22,24]. However, the correlation of miR-552 expression and its clinicopathological significance in CRC patients has not been investigated much.
This study aimed to verify the diagnostic and prognostic roles of miR-552 expression in patients with CRC. In this study, miR552 expression was investigated via quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) and analyzed for an association with clinicopathological features. Moreover, immunohistochemistry (IHC) for p53 and PTEN was conducted to confirm the association with miR-552.
A total of 80 patients with CRC were enrolled for this study. Normal mucosal tissues and primary cancer tissues were collected from the CRC patients who underwent surgical resection at Seoul National University Bundang Hospital. The histopathological differentiation was classified according to the World Health Organization standards (5th edition) [25], and pTNM was classified according to the American Joint Committee on Cancer (8th edition) [26]. The clinicopathological characteristics of each patient including sex, age, location, tumor size, and presence of lymphatic invasion were obtained from medical charts and pathology reports. The patients were followed up from the date of surgery either to the date of death or to the date of last follow-up.
The specimens of patients with CRC who underwent surgical resection were collected from the formalin-fixed-paraffin-embedded (FFPE) tissue. In each case, a 2 mm core sample was obtained from the tumor area, and the samples from 80 patients were arranged into two tissue microarray (TMA) blocks.
The RecoverALL Total Nucleic Acid Isolation Kit (Life Technologies, Carlsbad, CA, USA) was used to extract total RNA from the FFPE tissues. RNA extraction was performed as per the manufacturer’s manual. The purity and quantity of the extracted RNA were measured using a NanoDrop 2000/2000c spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA).
cDNA synthesis was performed on 10 ng of RNA from each sample using the TaqMan MicroRNA Reverse Transcript Kit (Applied Biosystems, Foster City, CA, USA). cDNA was synthesized using a C1000 Touch Thermal Cycler (Bio-Rad, Laboratories, Foster City, CA, USA) with the following cycle: 16°C for 30 minutes, 42°C for 30 minutes, and 85°C for 5 minutes.
The TaqMan small RNA assay kit (ABI, Applied Biosystems) was used to quantify miR-552, following the manufacturer’s instruction. qRT-PCR was conducted on the synthesized cDNA, to evaluate miRNA expression, with the following cycle: 95°C for 10 minutes, 95°C for 15 seconds, and 60°C for 1 minute. U6 small nuclear RNA was used as the standardized control for the target miRNA, miR-552.
qRT-PCR was performed to obtain the average value for quantifying the expression. Each value of the threshold cycle value (Ct value), obtained based on U6 expression was standardized and analyzed by a relative quantification method. U6 was used as the internal miRNA control, and the ∆Ct (to compare normal and tumor tissues) and ∆∆Ct values were calculated. The fold change of the gene was calculated using the estimated 2-∆∆Ct [27].
IHC for p53 (1:1,000, clone DO-7, DAKO, Santa Clara, CA, USA) and PTEN (1:1,000, clone Y184, Abcam, Cambridge, UK) was performed on 3-μm-thick sections from the TMA block using a Ventana automated immunostainer (BenchMark XT, Ventana Medical Systems, Tucson, AZ, USA), in accordance with the manufacturer's instructions. IHC was interpreted by intensity (negative, weak, and strong) and area (%). The PTEN and p53 expression were classified into three categories: (1) strong staining in ≥10% of the tumor cells was considered as strong positive, (2) samples without any nuclear staining of tumor cells were interpreted as negative, and (3) samples exhibiting weak staining in any area or patchy strong staining in <10% of tumor cells were regarded as weakly positive [28].
The miR-552 expression change was indicated by the median with range. High and low miR-552 subgroups were classified according to miR-552 expression levels with a cut-off of the median value [29]. Wilcoxon matched-pairs signed rank test was used to compare the miR-552 manifestations of the two groups, and the Kruskal-Wallis test was used to compare TNM stage sequential trends. For comparison of variables via category on clinical findings, the chi square or Fisher exact test was used. KaplanMeier curves with log-rank test were used for survival analysis. The Cox proportional hazards model was used for univariate and multivariate regression analyses. Linear-by-linear association analysis was used to compare miR-552 with p53, and PTEN expression patterns. PASW ver. 18.0 (IBM Co., Armonk, NY, USA) was used for statistical analysis. A p-value <.05 was considered statistically significant.
Primary cancer tissues and matched normal mucosal tissues were used to investigate the expression levels of miR-552 in patients with CRC. The relative expression value of miR-552 was identified based on the internal control of U6. The expression of miR-552 was approximately four times higher in primary cancer tissues than normal mucosal tissues; this was statistically significant (p<.001) (Fig. 1A).
qRT-PCR results showed that miR-552 expression increased in the primary cancer tissue of patients with CRC. Based on these results, the correlation with clinicopathological characteristics was analyzed by dividing the cases into low and high miR-552 subgroups based on the median of the miR-552 ∆∆Ct values (Table 1). Clinicopathological characteristics such as age, sex, tumor location, histologic grade, tumor size, pT, pN, M, pTNM stage, and venous invasion were compared with the miR-552 expression status. The miR-552 high expression group tended to be male, ocated in the right colon (ascending to transverse colon), have a low histologic grade, and be of advanced stage, but without statistical significance (p>.05).
When the median miR-552 values were compared according to pT and pTNM stage (Fig. 1B, C), the expression of miR-552 was higher in pT3–4 than pT1–2, and higher in stage II–IV than stage I, but the difference was not statistically significant (p=.393 and p=.414, respectively).
IHC was performed to determine the expression of p53 and PTEN proteins (Fig. 2) and the association with the miR-552 expression pattern was analyzed and compared. Negative expression of PTEN and p53 was more frequently found in the high miR-552 subgroup than in low subgroup (p=.068 and p=.004, respectively) (Table 2).
Our study showed that miR-552 was upregulated in cancer tissue when compared with normal tissue in patients with CRC. Thereafter, we performed univariate and multivariate survival analyses according to the miR-552 expression status. Kaplan-Meier survival curves showed that the miR-552 high subgroup was associated with worse prognosis, but this was not statistically significant (p=.255) (Fig. 3A). However, the miR-552 high and PTEN-negative subgroup had significantly worse outcome than the other subgroups (p=.029) (Fig. 3B). When multivariable Cox regression analysis was performed with pTNM stage and age, high miR-552 and PTEN-negative subgroup had a tendency of worse prognosis, but with borderline statistical significance (p=.077) (Supplementary Table S1).
The incidence of CRC is increasing every year. However, early diagnosis is difficult and the disease has a high mortality rate due to metastasis. Many recent studies have suggested altered expression of miRNAs in various cancers. For CRCs, researchers investigated how miRNAs contribute to carcinogenesis and regulate the invasion and metastasis of cancer during disease progression. miR-552, one of the miRNAs whose expression is altered in different types of cancers, has been found to be related to colorectal, ovarian, breast, liver, and pancreatic cancers [23,24,30-32]. In this study, the expression of miR-552 in normal mucosa and primary cancer tissue and the association with clinicopathological characteristics of patients with CRC were analyzed. The results showed that miR-552 in CRC showed an increased expression in primary cancer tissue compared to normal mucosal tissue. This is in accordance with the results of previous studies [19,33].
We determined the association between miR-552 and the tumor suppressor genes, PTEN and p53. PTEN catalyzes the dephosphorylation of phosphatidylinositol 3,4,5-trisphosphate (PIP3) [34]. Loss of PTEN function increases PIP3 activity and activates the phosphoinositide 3-kinase/protein kinase B (PKB or AKT; hereafter referred to as AKT) signaling pathway to promote cell growth, proliferation, and survival [35,36]. The protein phosphatase activity of PTEN is involved in regulating the cell cycle, preventing cells from growing or splitting too quickly [24,37]. p53 expression rapidly increases when DNA damage occurs in cells, leading to the expression of DNA repairing enzymes to allow cells to survive, or the expression of BCL-2–associated X protein and p53 upregulated modulator of apoptosis, which induce cell death, causing damaged cells to die on their own [22,38,39]. Abnormalities in PTEN and p53 expression promote cancer development by inducing cell cycle, reducing apoptosis, and decreasing genomic stability. Previous in vivo studies have shown that the 3'-untranslated region of PTEN and p53 mRNA has a potential miR-552 binding site and the two molecules interact, suggesting that miR-552 could regulate the expression of PTEN and p53 [22,24,40,41]. miR-552 directly targets PTEN and p53, reducing PTEN and p53 expression levels when miR-552 is overexpressed [22,24,40]. Our results also revealed frequent loss of PTEN expression or p53 negative expansion in the high miR-552 subgroup using clinical samples of CRC.
miR-552 had a tendency of increased expression in advanced pT or pTNM stage, but without statistical significance. In addition, the high miR-552 subgroup did not have a statistically significant worse prognosis. However, the high miR-552 subgroup with PTEN loss showed a statistically significant worse prognosis. It is suggested that the combined status of miRNAs and their target proteins may be more clinically significant than the miRNAs themselves. Further studies are needed to clarify it.
An analysis of the correlation between clinicopathological characteristics and miR-552 expression in patients with CRC showed that the expression of miR-552 was high regardless of age, sex, and tumor location (Table 1). However, a few previous studies have shown that high miR-552 expression in CRC and other cancers was correlated with aggressive clinicopathological factors, such as higher histologic grade, lymph node metastasis, and advanced TNM stage [19,33]. A potential reason why the results of our study differ from those of the previous studies could be the small sample size of our study. Therefore, a large multicenter study would be necessary in the future. In addition to cancer tissues of patients with CRC, miR-552 expression status in other tissues such as blood may be helpful for early diagnosis and detection of cancer recurrence.
miRNAs are expressed directly in cancer cells, or in cancer microenvironment such as cancer-associated fibroblasts (CAFs) and various tumor-infiltrating immune cells. miR-21 expression in CAFs was reported in CRC tissue using the in situ hybridization method [42]. Previous studies have reported that miR-552 expression increased in various cancer tissues and cell-lines including CRC, hepatocellular carcinoma, osteosarcoma, ovarian cancer, gastric cancer, pancreas cancer, and lung cancer, suggesting the possible expression of miR-552 in cancer cells. However, our results could not demonstrate which cells expressed miR-552 because we used the real-time PCR. Further studies are needed to reveal the source of miR-552 expression.
The results of this study suggest that increased miR-552 expression can be used as a diagnostic biomarker in patients with CRC and that the combined analysis with PTEN expression may be used as a prognostic marker, which can help diagnose and treat patients with CRC in the future.
Supplementary Information
The Data Supplement is available with this article at https://doi.org/10.4132/jptm.2021.01.17.
Notes
Ethics Statement
This study was by the Institutional Review Board of Seoul National University Bundang Hospital (B-1012/117-011). Informed consent was obtained from all individual participants included in the study
Author contributions
Conceptionalization: HSL. Funding Acquisition: HSL. Investigation: JI, SKN. Methodology: JI, SKN. Supervision: HSL. Writing—original draft: JI, HSL. Writing—review & editing: JI, SKN, HSL. Approval of final manuscript: all authors.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018; 68:394–424.

2. Hong S, Won YJ, Park YR, et al. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2017. Cancer Res Treat. 2020; 52:335–50.

3. Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009; 361:2449–60.
4. Luo X, Burwinkel B, Tao S, Brenner H. MicroRNA signatures: novel biomarker for colorectal cancer? Cancer Epidemiol Biomarkers Prev. 2011; 20:1272–86.

5. Koga Y, Yasunaga M, Takahashi A, et al. MicroRNA expression profiling of exfoliated colonocytes isolated from feces for colorectal cancer screening. Cancer Prev Res (Phila). 2010; 3:1435–42.

6. Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005; 37:495–500.

7. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993; 75:843–54.

8. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–97.
10. Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004; 101:2999–3004.

11. Ji J, Yamashita T, Wang XW. Wnt/beta-catenin signaling activates microRNA-181 expression in hepatocellular carcinoma. Cell Biosci. 2011; 1:4.

12. Kim NH, Kim HS, Kim NG, et al. p53 and microRNA-34 are suppressors of canonical Wnt signaling. Sci Signal. 2011; 4:ra71.

13. Cai J, Guan H, Fang L, et al. MicroRNA-374a activates Wnt/beta-catenin signaling to promote breast cancer metastasis. J Clin Invest. 2013; 123:566–79.
15. Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005; 435:834–8.

16. Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006; 103:2257–61.

17. Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006; 66:7390–4.

18. Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014; 20:460–9.

19. Wang N, Liu W. Increased expression of miR-552 acts as a potential predictor biomarker for poor prognosis of colorectal cancer. Eur Rev Med Pharmacol Sci. 2018; 22:412–6.
20. Xia ZS, Wang L, Yu T, et al. MiR-5000-3p, miR-5009-3P and miR-552: potential microRNA biomarkers of side population cells in colon cancer. Oncol Rep. 2014; 32:589–96.

21. Oberg AL, French AJ, Sarver AL, et al. miRNA expression in colon polyps provides evidence for a multihit model of colon cancer. PLoS One. 2011; 6:e20465.

22. Kwak B, Kim DU, Kim TO, Kim HS, Kim SW. MicroRNA-552 links Wnt signaling to p53 tumor suppressor in colorectal cancer. Int J Oncol. 2018; 53:1800–8.

23. Cao J, Yan XR, Liu T, et al. MicroRNA-552 promotes tumor cell proliferation and migration by directly targeting DACH1 via the Wnt/beta-catenin signaling pathway in colorectal cancer. Oncol Lett. 2017; 14:3795–802.
24. Zhao W, Han T, Li B, Ma Q, Yang P, Li H. miR-552 promotes ovarian cancer progression by regulating PTEN pathway. J Ovarian Res. 2019; 12:121.

25. Nagtegaal ID, Odze RD, Klimstra D, et al. The 2019 WHO classification of tumours of the digestive system. Histopathology. 2020; 76:182–8.

27. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008; 3:1101–8.

28. Hwang HJ, Nam SK, Park H, et al. Prediction of TP53 mutations by p53 immunohistochemistry and their prognostic significance in gastric cancer. J Pathol Transl Med. 2020; 54:378–86.
29. Tong F, Ying Y, Pan H, Zhao W, Li H, Zhan X. MicroRNA-466 (miR-466) functions as a tumor suppressor and prognostic factor in colorectal cancer (CRC). Bosn J Basic Med Sci. 2018; 18:252–9.

30. Kim HK, Lim NJ, Jang SG, Lee GK. miR-592 and miR-552 can distinguish between primary lung adenocarcinoma and colorectal cancer metastases in the lung. Anticancer Res. 2014; 34:2297–302.
31. Qu W, Wen X, Su K, Gou W. MiR-552 promotes the proliferation, migration and EMT of hepatocellular carcinoma cells by inhibiting AJAP1 expression. J Cell Mol Med. 2019; 23:1541–52.
32. Chen T, Lei S, Zeng Z, et al. Linc00261 inhibits metastasis and the WNT signaling pathway of pancreatic cancer by regulating a miR-5525p/FOXO3 axis. Oncol Rep. 2020; 43:930–42.

33. Li C, Wang Z, Chen S, Zhang J, Qu K, Liu C. MicroRNA-552 promotes hepatocellular carcinoma progression by downregulating WIF1. Int J Mol Med. 2018; 42:3309–17.

34. Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998; 273:13375–8.

35. Xiong B, Cheng Y, Ma L, Zhang C. MiR-21 regulates biological behavior through the PTEN/PI-3 K/Akt signaling pathway in human colorectal cancer cells. Int J Oncol. 2013; 42:219–28.

36. Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002; 2:489–501.
38. Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995; 80:293–9.

40. Han T, Zhang Y, Yang X, et al. miR-552 regulates liver tumor-initiating cell expansion and sorafenib resistance. Mol Ther Nucleic Acids. 2020; 19:1073–85.

41. Bi X, Lv X, Liu D, et al. METTL3-mediated maturation of miR126-5p promotes ovarian cancer progression via PTEN-mediated PI3K/Akt/mTOR pathway. Cancer Gene Ther 2020 Sep 16 [Epub]. https://10.1038/s41417-020-00222-3.

Fig. 1.
The expression level of miR-552 in the patients with colorectal cancer. (A) Comparative analysis of microRNA-552 (miR-552) expression in normal mucosal tissues (normal) and primary cancer tissues (tumor) in 80 patients with colorectal cancer using quantitative real-time reverse transcription polymerase chain reaction (by Wilcoxon matched-pairs signed rank test, ***p<.001). (B) miR-552 expression is relevant according to pT stages (Mann Whitney test; p=.393). (C) miR-552 expression is relevant according to pTNM stages (Kruskal-Wallis test, p=.414).
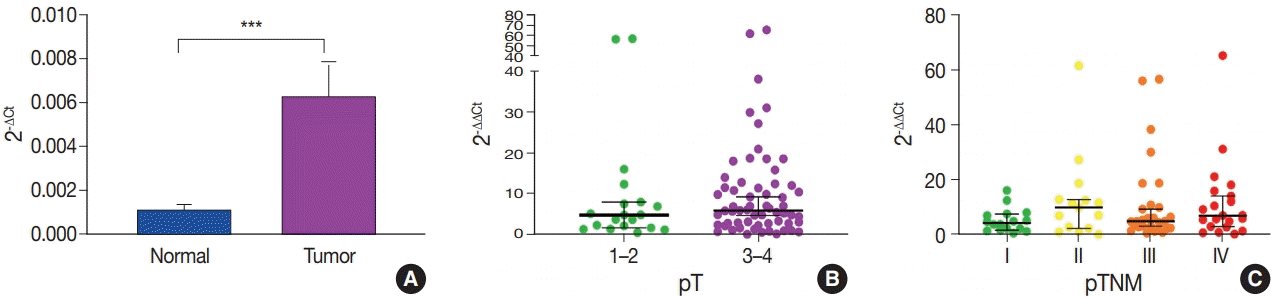
Fig. 2.
Immunohistochemical staining of phosphatase and tension homolog (PTEN) and p53 expression in patients with colorectal cancer: (A) negative staining of PTEN, (B) weak positive staining of PTEN, (C) positive staining of PTEN, (D) negative staining of p53, (E) weak positive staining of p53, and (F) positive staining of p53. (G) Representative amplification curves of microRNA-552 (miR-552) and U6 by real-time polymerase chain reaction (PCR) methods. NTC, no template control.
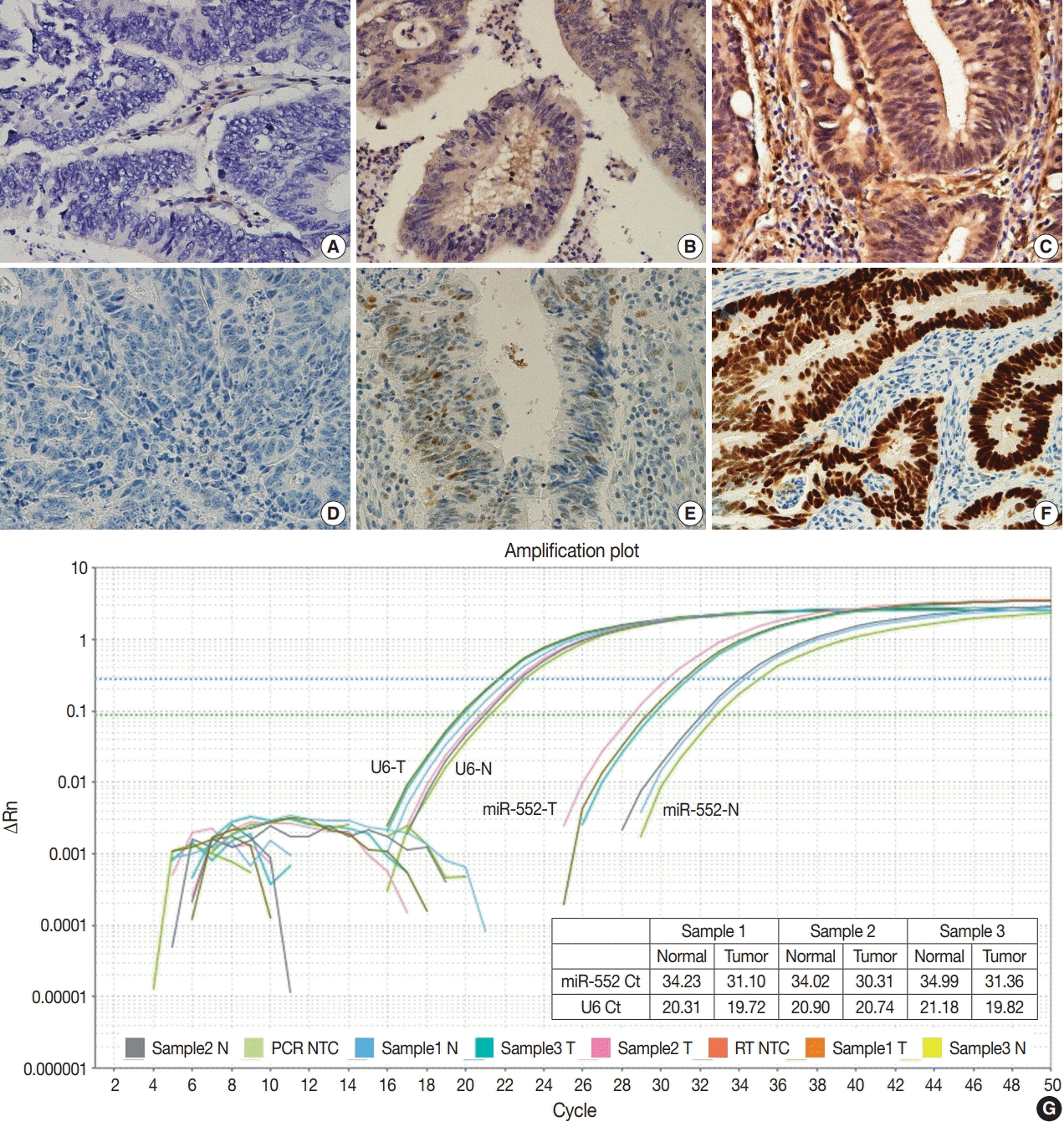
Fig. 3.
Kaplan-Meier univariate survival analysis according microRNA-552 (miR-552) and phosphatase and tension homolog (PTEN) expression status in patients with colorectal cancer. (A) miR-552 high group has worse prognosis than low group, but is not statistically significant (p=.255). (B) miR-552 high and PTEN-negative group is significantly associated with poor prognosis when compared to others (p=.029).
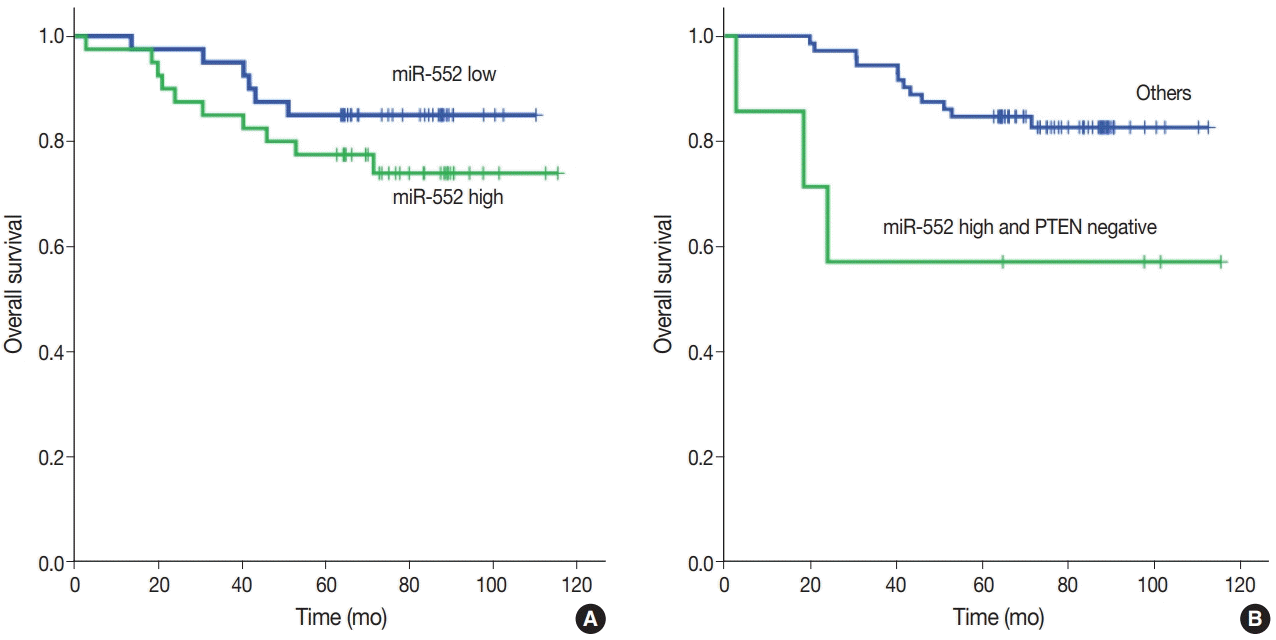
Table 1.
Correlation between the clinicopathological characteristics of patients with colorectal cancer and miR-552 expression
Table 2.
The relationship between PTEN and p53 protein expression by immunohistochemistry and miR-552 expression (linear-by-linear association)




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download