Abstract
Blood liquid biopsy has emerged as a way of overcoming the clinical limitations of repeat biopsy by testing for the presence of acquired resistance mutations to therapeutic agents. Despite its merits of repeatability and non-invasiveness, this method is currently only used as a supplemental test due to a relatively low sensitivity rate of 50%–60%, and cannot replace tissue biopsy. The circulating tumor DNAs used in blood liquid biopsies are passive products of fragmented DNA with a short half-life released following tumor cell death; the low sensitivity seen with liquid blood biopsy results from this instability, which makes increasing the sensitivity of this test fundamentally difficult. Extracellular vesicles (EVs) are ideal carriers of cancer biomarkers, as cancer cells secret an abundance of EVs, and the contents of tumor cell-originated EVs reflect the molecular and genetic composition of parental cells. In addition, EV-derived DNAs (EV DNAs) consist of large-sized genomic DNAs and tumor-specific oncogenic mutant DNAs. For these reasons, liquid biopsy using EV DNA has the potential to overcome issues arising from tissue shortages associated with small biopsies, which are often seen in lung cancer patients, and the biopsy product can be used in other diagnostic methods, such as epidermal growth factor receptor (EGFR) mutation testing and next-generation sequencing (NGS). A higher sensitivity can be achieved when EV DNAs obtained from bronchoalveolar lavage fluid (BALF) are used rather than those from blood. BALF, when obtained close to the tumor site, is a promising liquid biopsy tool, as it enables the gathering of both cellular and non-cellular fractions of the tumor microenvironment, and provides increased diagnostic sensitivity when compared to blood.
Go to : 
A growing trend in identifying biomarkers in diseases has brought on a dramatic rise in the number of tissue biopsies performed in the era of precision medicine. With advanced non-small cell lung cancer (NSCLC) patients often relying on small biopsies, the clinical need for an established liquid biopsy protocol has become an urgent necessity. Unlike gastrointestinal cancer, where tumor lesions in all locations can generally be visualized, lung cancers can only be approached by bronchoscopy, which limits visualization to central tumors. Even with the use of endobronchial ultrasound, peripheral lung cancers, such as adenocarcinoma before metastasis to the mediastinal lymph node, must rely on the invasive percutaneous needle biopsy (PCNB).
Recent trends have shown a decrease in the incidence of central lung cancers, such as squamous cell carcinoma and small cell carcinoma, with a concomitant increase in the incidence adenocarcinoma, where testing to identify driver oncogenic mutations is essentially useful [1-3]. With the development of target therapy drugs, the clinical need for rebiopsy or even repeated biopsies is increasing in order to identify drug resistance mechanisms in the targeted tissues [4,5]. Lung cancer tumor lesions are often small in size or positioned in a way that makes them difficult to target. In the case of ground-glass nodules (GGNs), a biopsy is not possible until enough of a solid portion develops to obtain tumor tissue by PCNB. For these reasons, “tissue is the issue” is a challenge faced by many clinicians, especially by those treating lung cancers. Liquid biopsy using blood was introduced to overcome the difficulties of obtaining a quality solid tumor tissue sample, but remains a secondary diagnostic technique to tissue biopsies due to its low sensitivity [6-8].
Blood liquid biopsy uses circulating tumor DNAs, free-floating DNA fragments found in the blood, has a low sensitivity, as the sample is not from near the tumor site and the DNA is instable due to its nakedness [9]. Since blood is a complex body fluid with many components, there is a need for a different biosource for use in liquid biopsy. Also, the characteristics of cell-free DNAs (cfDNAs) extracted from plasma after the removal of red blood cells have never been properly defined, as these DNAs could have originated from free-floating DNA, DNA-protein complexes, and/or inside of extracellular vesicles (EVs). Extracted cfDNAs are mostly found in fragments of about 180 bp in length, which, in the context of cancer patients, circumstantially indicates that they are passively released byproducts from tumor cell death. The free-floating nature gives cfDNAs a short half-life of 2–2.5 hours, and this instability makes them inadequate as biomarkers [10,11].
EVs, nano-sized vesicles secreted by almost all types of cells, carry bioactive molecules, such as proteins, glycans, lipids, metabolites, RNAs, and DNAs, enclosed by a lipid bilayer, and act as an essential mediator in cell-to-cell communication [12-15]. EVs make an ideal cancer biomarker, as the contents of tumor celloriginated EVs reflect the molecular and genetic composition of parental cells, and are secreted in higher amounts than EVs of normal cells [16]. DNAs inside of an EV have advantages over cfDNAs as a biosource for liquid biopsy. Foremost, EV-derived DNAs (EV DNAs) consist of large-sized genomic DNAs and tumor-specific oncogenic mutant DNAs, unlike the fragmented cfDNAs [17]. However, EVs have yet to be precisely defined and categorized. For the purpose of this review, our main focus will be to classify the exosomes found in EVs and recapitulate the current status of liquid biopsy in lung cancer, focusing on EVs.
Most cancers, even when they remain localized to a single primary site, can have a systemic impact by releasing tumor cells and byproducts into various body fluids. Liquid biopsy makes use of these biofluids, such as blood, urine, cerebrospinal fluid, pleural effusion, and glandular secretions, to analyze the characteristics of tumor cells in a continuous and noninvasive matter [18,19]. There are four major biosources used for liquid biopsy: circulating tumor cells (CTCs), cfDNAs, tumor-educated platelets (TEPs), and EVs [20].
CTCs are shed by primary tumors and stay circulating in the bloodstream until they metastasize to various parts of the body, which can happen at any time, even in the early stages of cancer development [21,22]. The advantage of using CTCs over other biosources is that a multi-omics approach evaluating the genomic, transcriptomic, and proteomic profiles of the tumor can be applied [23]. However, due to their minimal presence (there are approximately 1–10 CTCs/mL in blood), identification and characterization of cancer using CTCs is proving to be difficult [24,25], while the heterogeneity of surface markers and size of CTCs makes them challenging for clinical use [22]. The isolation of CTCs is also quite complicated, due to their extreme rarity compared to surrounding blood cells. The CellSearch system is the only U.S. Food and Drug Administration (FDA)–approved commercial product for CTC enumeration, but it is not widely used in a clinical setting because of its shortcomings, such as its high cost, need for manual processing, and high false-positive/false-negative rates [26]. Microfluidic platforms that are affinity-based and that utilize surface markers such as epithelial cell adhesion molecule to distinguish CTCs from surrounding blood cells have been proposed, but the matter of a high false-negative rate remains challenging due to the downregulation of surface epithelial markers driven by the epithelial-mesenchymal transition (EMT) [27,28]. Despite extensive research, CTCs are currently not commonly used in NSCLC mainly due to technical problems.
CfDNAs are the most commonly used biosource for liquid biopsy in lung cancer, because they are easily obtained and their concentration is significantly higher in cancer patients compared to healthy persons, with this increase correlating with cancer stage [22,29,30]. The Cobas EGFR mutation test v2 is clinically used for detecting T790M resistance mutation and in the prescription of osimertinib [31]. In recent years, next-generation sequencing (NGS) using blood-derived cfDNAs has been applied for screening and early diagnosis, treatment selection and prognosis, and residual disease and risk of relapse in NSCLC patients [32]. We are not going to review this area deeply, as many research articles and reviews have already explored blood cfDNA liquid biopsy, especially in epidermal growth factor receptor (EGFR)-mutated NSCLC patients [32-36]. The most important thing in blood cfDNA liquid biopsy in NSCLC is that cfDNA has fundamental limitations for liquid biopsy due to the fragmented nature of cfDNA and its instability, which leads to low test sensitivity.
TEP was recently discovered as a potential noninvasive biomarker, as TEPs are involved in the initiation, progression, and metastasis of tumors [37,38]. Cancer cells induce platelet activation and assist the production of TEPs, which can promote tumor cell invasion through regulation of the p38MAPK-MMP9 pathway, promoting metastasis through EMT and escaping immunity via platelets-coated CTCs [39]. Also, alteration in a panel of RNAs was observed in cancers, which could also be useful as a biomarker [40]. Best et al. [41] reported the development of a TEP algorithm that can predict the presence of EGFR mutations, MET amplification, and KRAS mutations with high accuracy, which could potentially be used in the diagnosis of NSCLC.
Go to : 
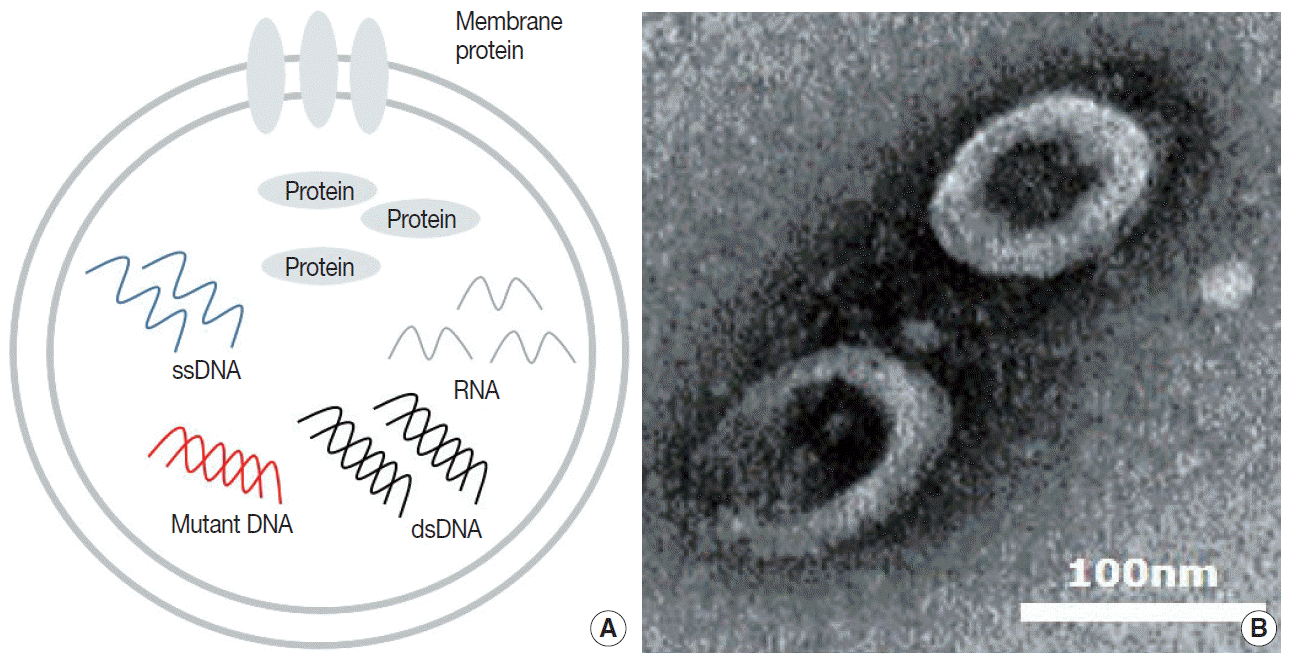
Tumor cells actively produce and release EVs that carry cytoplasmic components, including RNAs, DNAs, and proteins (Fig. 1A) [15,42]. These tumor-derived EVs are found in all bodily fluids, including saliva, blood, urine, and bronchoalveolar lavage fluid (BALF) (Fig. 1B) [43,44]. Mechanisms through which tumor-derived EVs select their cargo, serve the tumor, and affect surrounding cells are under intense investigation. There are multiple hypotheses for the original function of EVs in the context of cancer, from containing toxin and quorum sensing molecules in bacterial EVs to carrying information and conveying genetic messages by the tumor cells [45-47].
Many reports have illustrated that EVs derived from tumors play a significant role in intercellular communication by transmitting signals and transferring their oncogenic contents. These roles include the promotion of oncogenic potential in acceptor cells by increasing their cell proliferation potential [48], modulation of metastatic ability [49], and up-regulation of angiogenesis [50]. Studies have demonstrated that EVs are sufficient to recapitulate the formation of tumors through the neoplastic reprogramming of stem cells using prostate cancer cell-derived EVs [51], and breast cancer cell-derived EVs can promote normal epithelial cells to form tumors in a Dicer-dependent manner [52]. Recent studies have shown that cancer cells secrete immunologically active EVs which mediate immune regulation in the tumor microenvironment (TME) by acting as mediators of intercellular communication [53]. In lung cancers, EVs are shown to act as mediators of intercellular communication and play a vital role in tumor growth, progression, invasion, metastasis, and targeted drug resistance [54-57]. Furthermore, as EVs contain the bioactive molecules of the cells they are derived from, they have great potential as diagnostic biomarkers [14,15,17].
Go to : 
In the era of personalized medicine, liquid biopsy is becoming an indispensable method for dealing with medical issues, such as the early detection of diseases, improving the accuracy of diagnosis, predicting responses of patients to drugs, and selecting follow-up of treatments [18,19]. In this sense, the discovery of target bio-molecules that can be used as biomarkers is as important as the advancement of technology used for liquid biopsy. The EVs secreted by cancer cells, enriched with various proteins, DNAs, and RNAs, have diagnostic value as biomarkers for clinical applications [12,13]. It is well known that EVs carry various types of proteins and RNAs from the cells that originally secreted them, which make them particularly fit as biomarkers for human disease, especially cancer. Many different types of protein, including membrane and cytosolic proteins, involved in various biological functions have been identified in EVs.
In recent years, an increasing number of studies have described the possible use of EV-derived proteins and RNAs as biomarkers. For example, Logozzi et al. [58] developed a method to detect and quantify a tumor-associated marker, a membrane protein called caveolin-1, from EVs. Recent identification of distinct integrin expression patterns in different types of cancer, including through the use of EV proteomics in lung cancer [59], measuring delta-catenin in prostate cancer-derived EVs [60], measuring DEL-1 levels from plasma EVs for early detection of breast cancer [61] and using proteomic profiling to identify subtype-specific protein clusters in breast cancer-derived EVs [62], has revealed the potential to use these expression patterns as cancer detection biomarkers. Many studies have identified elevation or downregulation of specific miRNAs in cancer patients that could be used as markers in the prognosis of NSCLC [63,64]. EV DNAs have been identified in both the double-stranded genomic and mutated fragmented forms [14,16,65].
It is commonly accepted that EV cargo packaging is not a random process, but rather a selective process that represents the cell of origin, which makes EVs and their contents a good representation of cancer. Also, EVs have a long half-life and are well protected in the human body by having an outer membrane. All of these studies indicate that EVs can be used as noninvasive biomarkers. Moreover, from the perspective of liquid biopsy, EVs can offer many advantages.
Go to : 
Unlike the relatively well-characterized proteins and RNAs found in EVs, until recently EV DNAs have not been the center of attention among researchers. Thakur et al. [65] identified tumor-derived EVs carrying double-stranded DNA (dsDNA) and EV DNA that represented the entire genome, including reflecting the mutational status, of parental tumor cells. Several studies also have identified that while EV DNAs exist in short and long sizes, most are 10 kb or longer, and have demonstrated that detection of cancer-related mutations can be done sufficiently with EV DNAs [16,66,67]. A study characterized subpopulations within the EV population with different DNA and RNA content and topology. In addition, they identified two types of DNAs, internally protected and external surface-related DNA, that could be used as a basis for developing diagnostic methods [68,69].
The fragmented nature of cfDNAs makes it difficult to generate a reliable genomic characterization for NGS, which in turn requires barcode and deep sequencing [70,71]. Contrarily, long-stranded and concentrated EV DNA is easy to amplify and therefore makes it a suitable candidate for NGS. In short, the use of EV DNA for NGS requires the same processes as those required in the well-established methods which use a tissue sample [72]. It has been suggested that NGS using EVs isolated from pleural effusions and urine could effectively replace tissue-based NGS in cases where there is a shortage of tissue [73,74]. EV DNAs have several important differences from the more clinically established cfDNA, as shown in Table 1. The encapsulation of DNA within the lipid bilayer of EVs seems to enhance the stability of the DNA by shielding it from the outer environment, unlike the ‘naked’ cfDNA, which is directly exposed to bodily fluids. The amount of cfDNA released by tumors is normally low, and represents only the fraction of the tumor genomic heterogeneity released by dying tumor cells. By contrast, the process for isolating EVs from fluids increases the final concentration of EV DNAs obtained, and EVs are secreted by both viable and dying tumor cells [44,75]. In a study comparing the use of plasma EV DNAs and cfDNAs of NSCLC patients in liquid biopsies, EV DNA improved the detection sensitivity in EGFR profiling of the exon 19 deletion, L858R, and T790M mutations. This study also highlighted the ongoing diagnostic and prognostic ability of EV DNA by showing that the mutations identifiable by EV DNA liquid biopsy included the acquired resistance specific mutation T790M. Furthermore, the specificity and sensitivity of liquid biopsy increased even more with the use of BALF EV DNA [16,76]. Similar to NSCLC, analysis of serum EV DNA from pancreatic cancer patients also demonstrated the presence of clinically relevant KRAS-specific mutations [14,77,78].
Comparison of EV DNA and cfDNA
As previously described, EVs are widely distributed in various body fluids, which make them more easily accessible with less invasive methods than tumor cells. More importantly, the specificity and sensitivity can be significantly increased when EV DNAs are used for liquid biopsy compared to cfDNAs. For these reasons, liquid biopsy of EV DNAs could be a clinically useful means of isolating cancer-specific DNA, while achieving a higher sensitivity and stability than the current diagnostic methods.
Go to : 
Compared to the relatively invasive and expensive tissue biopsies, liquid biopsies using blood better allows for repeatable evaluation of the disease status of cancer patients, making it an especially useful tool in clinical practice. Recently, many have studied liquid biopsy using cfDNA, which is released into blood through cell death mechanisms, such as necrosis and apoptosis, has a short half-life, and is detected at very low concentrations (5–10 ng/mL). Because of these characteristics, high-sensitivity approaches are needed when using cfDNA. For example, the FDA-approved Cobas tool for detecting mutations with plasma cfDNA has a 69%– 86% sensitivity for EGFR mutations and a 55–75% sensitivity for T790M mutation [79-81]. Also, the mutation detection rate in intrathoracic metastatic diseases (M1a) is much lower (39% for activation mutations, 27% for T790M) [82]. To overcome this low sensitivity, various high technology platforms, such as droplet digital polymerase chain reaction (PCR) [81] or BEAMing (Beads, Emulsion, Amplification, and Magnetics) technology, were introduced by several research institutes [79,82]. Briefly, the BEAMing assay uses individual DNA molecules that are attached to magnetic beads in water-in-oil emulsions and processed by compartmentalized PCR amplification. However, the cost of these ultra-fast sequencing technologies is too high for use in most clinics.
EV miRNAs have been shown to be promising biomarkers for the diagnosis of NSCLC, because EVs have a large amount of tumor-derived RNA [83], and RNAs within EVs are resistant to the activity of external RNases. MiRNAs are short, non-coding, single-stranded RNA molecules (19–22 nucleotides) that target complementary mRNA sequences and play important roles in regulating gene expression at the post-transcriptional level. For example, miR-660-5p [84], miR-17-5p [85], miR-126 [86], and a multiple miRNAs combination panel [87] are expressed at higher levels in NSCLC patients compared to healthy controls. MiRNAs from circulating EVs seem to be associated with the stage, tumor grade, histology, and prognosis of cancer patients [87,88], but the sensitivity and specificity of these connections remained at only 60%–80%.
The analysis of EV nucleic acids in plasma showed that they are more sensitive in identifying relevant mutations than cfDNAs. Krug et al. [89] showed the EV RNA was significantly more sensitive in detecting activating EGFR mutations (98%) and T790M mutation (90%) compared to the matched cfDNA (82% for activating EGFR mutations, 84% for T790M). The RNAs from the captured EVs are reverse transcribed to cDNAs, and subsequently analyzed by targeted NGS. These methods increase the sensitivity of detecting EGFR mutations but require the high-cost NGS assay. A number of studies on liquid biopsy using EVs isolated from blood have been performed, but the sensitivity of this assay reported in these studies was not yet high enough to replace tissue biopsy. Thus, more sensitive assays must be developed.
Go to : 
Malignant pleural effusions are a clinical problem in up to one-third of patients with advanced NSCLC [90,91]. Liquid biopsy of malignant pleural effusions has a relatively low sensitivity for the diagnosis of NSCLC, at approximately 60% [92]. For EGFR genotyping, cellular components, including cell block or cytology, are conventionally used, while supernatant fractions are usually discarded. Blind or closed needle pleural biopsy is sometimes indicated, although the diagnostic yield is typically low. Pleuroscopy, also known as medical thoracoscopy, has a higher diagnostic yield and provides better diagnostic sensitivity, but it is a highly invasive procedure. Even with pleuroscopy, adequate tissue biopsies are frequently unavailable due to co-morbidities or for other reasons.
A recent study demonstrated the superiority of using the cell-free supernatant from pleural effusions for EGFR mutation detection over using cell pellets [93-95]. EGFR genotyping using EV DNAs and cfDNAs from the supernatant of pleural effusions resulted in 100% agreement with tissue EGFR genotyping in both EGFR–tyrosine kinase inhibitor (TKI) naïve patients and patients who had acquired resistance to EGFR-TKI [96]. In addition, it showed that the T790M detection rate using EV DNAs as a biosource was superior to using cell blocks or cfDNAs. These results suggest that the supernatant of pleural effusions is particularly effective for EGFR genotyping for patients with pulmonary adenocarcinoma when compared to doing conventional cytology or using cell block samples. Therefore, using EV DNAs from the supernatant of pleural effusions is promising for EGFR genotyping, including T790M detection, in pulmonary adenocarcinoma patients who develop pleural effusions.
Go to : 
Bronchoalveolar lavage (BAL) is a procedure primarily done for diagnostic purposes by which the cellular and non-cellular contents of bronchial and alveolar spaces are obtained [97]. The advantages of BAL are that it is a minimally invasive method and provides access to the disease-located site of diffuse interstitial lung diseases. For lung cancer, BAL can provide a unique gateway to access the TME, where tumor cells are known to shed an abundance of EVs [98]. Correspondingly, the results of liquid biopsy using EVs isolated from the BALF of NSCLC patients have been shown to be superior to liquid biopsy using plasma cfDNAs.
Recent studies have demonstrated that EVs successfully isolated from the BALF of lung cancer patients contain an abundant amount of dsDNAs, and that liquid biopsy for EGFR genotyping using BALF is tissue-specific and extremely sensitive compared to using cfDNAs [16,76]. In a study performed with pathologically confirmed NSCLC patients, the sensitivity and specificity of BALF EV-based EGFR genotyping were high, and this assay showed an even better mutation detection rate than tissue/cytology-based typing. Also, the sensitivity increased as the stage progressed, reaching 100% with stage IV patient samples [76]. These results seem to suggest BALF EV-based EGFR genotyping is a highly promising liquid biopsy method, and particularly efficient for patients that require repeat biopsies throughout progression of the disease.
Go to : 
Tissue shortages caused by small biopsy sizes is emerging as a hurdle to the adequate treatment of advanced NSCLC patients in the era of precision oncology. A variety of molecular, genetic, and/or genomic tests, including real-time PCR, reverse transcription polymerase chain reaction, fluorescence in situ hybridization, immunohistochemistry, and NGS, are performed to match the right drugs to the right patients. A second or even third biopsy is frequently performed to investigate the acquired resistance to targeted therapeutic agents, such as EGFR or T790M mutations. The incidence of peripheral lung cancers, such as adenocarcinoma, which is usually confirmed by PCNB, is increasing. Moreover, the location of lung cancer development is clearly moving from central airways, in the past, to small airways or alveoli at present. GGNs detected by low-dose computed tomography are typically not accessible without surgical resection. Lung biopsy is an indispensable procedure for the diagnosis and treatment of lung cancer patients, but it is difficult to obtain adequate tissue for precision oncology in lung cancer. All of these are essential reasons for adopting liquid biopsy rapidly into the diagnosis and treatment of lung cancer, especially for NSCLC. As mentioned above, although cfDNA has been popularly used, it has intrinsic defects as a biosource for liquid biopsy due to its low stability. Although the use of improved technology with superior sensitivities, like digital PCR, BEAMing, and ultra-deep sequencing, can improve its utility to some degree, blood liquid biopsy using cfDNA cannot ultimately replace tissue biopsy due to its shortcomings. Specifically, that cfDNA is passively released from dead or dying tumor cells is a critical limitation, while EV DNA is actively released by living tumor cells, thereby reflecting their characteristics in real time.
Liquid biopsy will likely face challenges to becoming a reliable biosource despite technological advances. In this regard, EVbased BALF liquid biopsy is a highly-anticipated liquid biopsy platform for lung cancers. In the light of recent increases in the occurrence of peripheral lung adenocarcinomas, which carry a high risk of complications, the use of BALF is advantageous as it provides a unique, direct access route to the TME and a way to collect tumor-derived EVs in vivo. Studies on EGFR genotyping using BALF EV DNAs demonstrated that the use of EV DNAs, which are representative of genomic DNA, could yield results equivalent to those obtained using tissue DNA.
EV-based liquid biopsies are not widely available in hospitals and medical labs at present, as they require specific technology and equipment. For example, multiple different methods are being used to isolate EVs, such as differential centrifugation or density gradients used to remove larger cellular debris and isolate EVs, which means the biological properties and contents of the EVs are largely influenced by the method of isolation. Therefore, the optimization and validation of EV isolation methods remains crucial to establish EVs as a biomarker for diagnostic use. Nevertheless, with its high sensitivity and specificity, the application of liquid biopsy using EV DNAs is expected to become more widely used clinically in the near future when an established method becomes available.
Go to : 
Notes
Author contributions
Conceptualization: KYL, IAK, JYH. Investigation: IAK, JYH, SEL, WSK. Supervision: KYL. Writing—original draft: KYL, IAK, JYH, HJK. Writing—review & editing: KYL, IAK, JYH. Approval of final manuscript: all authors.
Go to : 
References
1. Barta JA, Powell CA, Wisnivesky JP. Global epidemiology of lung cancer. Ann Glob Health. 2019; 85:8.

2. de Groot PM, Wu CC, Carter BW, Munden RF. The epidemiology of lung cancer. Transl Lung Cancer Res. 2018; 7:220–33.

3. Dela Cruz CS, Tanoue LT, Matthay RA. Lung cancer: epidemiology, etiology, and prevention. Clin Chest Med. 2011; 32:605–44.

4. Wu SG, Shih JY. Management of acquired resistance to EGFR TKI-targeted therapy in advanced non-small cell lung cancer. Mol Cancer. 2018; 17:38.

5. Imakita T, Matsumoto H, Hirano K, Morisawa T, Sakurai A, Kataoka Y. Impact on prognosis of rebiopsy in advanced non-small cell lung cancer patients after epidermal growth factor receptor-tyrosine kinase inhibitor treatment: a systematic review. BMC Cancer. 2019; 19:105.

6. Bollinger MK, Agnew AS, Mascara GP. Osimertinib: A third-generation tyrosine kinase inhibitor for treatment of epidermal growth factor receptor-mutated non-small cell lung cancer with the acquired Thr790Met mutation. J Oncol Pharm Pract. 2018; 24:379–88.

7. Akamatsu H, Katakami N, Okamoto I, et al. Osimertinib in Japanese patients with EGFR T790M mutation-positive advanced non-small-cell lung cancer: AURA3 trial. Cancer Sci. 2018; 109:1930–8.

8. Odogwu L, Mathieu L, Goldberg KB, et al. FDA benefit-risk assessment of osimertinib for the treatment of metastatic non-small cell lung cancer harboring epidermal growth factor receptor T790M mutation. Oncologist. 2018; 23:353–9.

9. Cheng F, Su L, Qian C. Circulating tumor DNA: a promising biomarker in the liquid biopsy of cancer. Oncotarget. 2016; 7:48832–41.

10. Mouliere F, Chandrananda D, Piskorz AM, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med. 2018; 10:eaat4921.

11. Gorgannezhad L, Umer M, Islam MN, Nguyen NT, Shiddiky MJ. Circulating tumor DNA and liquid biopsy: opportunities, challenges, and recent advances in detection technologies. Lab Chip. 2018; 18:1174–96.

12. Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014; 30:255–89.

13. Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002; 2:569–79.

14. Kahlert C, Melo SA, Protopopov A, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014; 289:3869–75.

15. Armstrong D, Wildman DE. Extracellular vesicles and the promise of continuous liquid biopsies. J Pathol Transl Med. 2018; 52:1–8.

16. Hur JY, Kim HJ, Lee JS, et al. Extracellular vesicle-derived DNA for performing EGFR genotyping of NSCLC patients. Mol Cancer. 2018; 17:15.

17. Cui S, Cheng Z, Qin W, Jiang L. Exosomes as a liquid biopsy for lung cancer. Lung Cancer. 2018; 116:46–54.

18. Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013; 10:472–84.

19. Pantel K, Alix-Panabieres C. Liquid biopsy and minimal residual disease: latest advances and implications for cure. Nat Rev Clin Oncol. 2019; 16:409–24.
20. Bracht JW, Mayo-de-Las-Casas C, Berenguer J, Karachaliou N, Rosell R. The present and future of liquid biopsies in non-small cell lung cancer: combining four biosources for diagnosis, prognosis, prediction, and disease monitoring. Curr Oncol Rep. 2018; 20:70.

21. Rolfo C, Castiglia M, Hong D, et al. Liquid biopsies in lung cancer: the new ambrosia of researchers. Biochim Biophys Acta. 2014; 1846:539–46.

22. Calabuig-Farinas S, Jantus-Lewintre E, Herreros-Pomares A, Camps C. Circulating tumor cells versus circulating tumor DNA in lung cancer-which one will win? Transl Lung Cancer Res. 2016; 5:466–82.
23. Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014; 4:650–61.

24. Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004; 351:781–91.

25. Krebs MG, Hou JM, Sloane R, et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J Thorac Oncol. 2012; 7:306–15.

26. Poudineh M, Sargent EH, Pantel K, Kelley SO. Profiling circulating tumour cells and other biomarkers of invasive cancers. Nat Biomed Eng. 2018; 2:72–84.

27. Nagrath S, Sequist LV, Maheswaran S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007; 450:1235–9.

28. Qian W, Zhang Y, Chen W. Capturing cancer: emerging microfluidic technologies for the capture and characterization of circulating tumor cells. Small. 2015; 11:3850–72.

29. Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014; 32:579–86.

30. Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977; 37:646–50.
31. Karachaliou N, Sosa AE, Molina MA, Centelles Ruiz M, Rosell R. Possible application of circulating free tumor DNA in non-small cell lung cancer patients. J Thorac Dis. 2017; 9(Suppl 13):S1364–72.

32. Chen M, Zhao H. Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum Genomics. 2019; 13:34.

33. Goldman JW, Noor ZS, Remon J, Besse B, Rosenfeld N. Are liquid biopsies a surrogate for tissue EGFR testing? Ann Oncol. 2018; 29:i38–46.

34. Hochmair MJ, Buder A, Schwab S, et al. Liquid-biopsy-based identification of EGFR T790M mutation-mediated resistance to afatinib treatment in patients with advanced EGFR mutation-positive NSCLC, and subsequent response to osimertinib. Target Oncol. 2019; 14:75–83.
35. Rolfo C, Mack PC, Scagliotti GV, et al. Liquid biopsy for advanced non-small cell lung cancer (NSCLC): a statement paper from the IASLC. J Thorac Oncol. 2018; 13:1248–68.
36. Wan JC, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017; 17:223–38.

37. Best MG, Wesseling P, Wurdinger T. Tumor-educated platelets as a noninvasive biomarker source for cancer detection and progression monitoring. Cancer Res. 2018; 78:3407–12.

38. Nilsson RJ, Karachaliou N, Berenguer J, et al. Rearranged EML4-ALK fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancer. Oncotarget. 2016; 7:1066–75.

39. Liu L, Lin F, Ma X, Chen Z, Yu J. Tumor-educated platelet as liquid biopsy in lung cancer patients. Crit Rev Oncol Hematol. 2020; 146:102863.

40. McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol. 2014; 16:717–27.

41. Best MG, Sol N, Kooi I, et al. RNA-Seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell. 2015; 28:666–76.

42. Choi DS, Kim DK, Kim YK, Gho YS. Proteomics of extracellular vesicles: exosomes and ectosomes. Mass Spectrom Rev. 2015; 34:474–90.

43. Keller S, Ridinger J, Rupp AK, Janssen JW, Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J Transl Med. 2011; 9:86.

44. Carnino JM, Lee H, Jin Y. Isolation and characterization of extracellular vesicles from broncho-alveolar lavage fluid: a review and comparison of different methods. Respir Res. 2019; 20:240.

45. Aldick T, Bielaszewska M, Uhlin BE, Humpf HU, Wai SN, Karch H. Vesicular stabilization and activity augmentation of enterohaemorrhagic Escherichia coli haemolysin. Mol Microbiol. 2009; 71:1496–508.
46. Dutta S, Iida K, Takade A, Meno Y, Nair GB, Yoshida S. Release of Shiga toxin by membrane vesicles in Shigella dysenteriae serotype 1 strains and in vitro effects of antimicrobials on toxin production and release. Microbiol Immunol. 2004; 48:965–9.
47. Choi D, Lee TH, Spinelli C, Chennakrishnaiah S, D'Asti E, Rak J. Extracellular vesicle communication pathways as regulatory targets of oncogenic transformation. Semin Cell Dev Biol. 2017; 67:11–22.

48. Soldevilla B, Rodriguez M, San Millan C, et al. Tumor-derived exosomes are enriched in DeltaNp73, which promotes oncogenic potential in acceptor cells and correlates with patient survival. Hum Mol Genet. 2014; 23:467–78.
49. Peinado H, Aleckovic M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012; 18:883–91.
50. Umezu T, Ohyashiki K, Kuroda M, Ohyashiki JH. Leukemia cell to endothelial cell communication via exosomal miRNAs. Oncogene. 2013; 32:2747–55.

51. Abd Elmageed ZY, Yang Y, Thomas R, et al. Neoplastic reprogramming of patient-derived adipose stem cells by prostate cancer cell-associated exosomes. Stem Cells. 2014; 32:983–97.

52. Melo SA, Sugimoto H, O'Connell JT, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014; 26:707–21.

53. Greening DW, Gopal SK, Xu R, Simpson RJ, Chen W. Exosomes and their roles in immune regulation and cancer. Semin Cell Dev Biol. 2015; 40:72–81.

54. Hsu YL, Hung JY, Chang WA, et al. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene. 2017; 36:4929–42.

55. Li X, Wang S, Zhu R, Li H, Han Q, Zhao RC. Lung tumor exosomes induce a pro-inflammatory phenotype in mesenchymal stem cells via NFkappaB-TLR signaling pathway. J Hematol Oncol. 2016; 9:42.

56. Fujita Y, Kosaka N, Araya J, Kuwano K, Ochiya T. Extracellular vesicles in lung microenvironment and pathogenesis. Trends Mol Med. 2015; 21:533–42.

57. Jung JH, Lee MY, Choi DY, et al. Phospholipids of tumor extracellular vesicles stratify gefitinib-resistant nonsmall cell lung cancer cells from gefitinib-sensitive cells. Proteomics. 2015; 15:824–35.

58. Logozzi M, De Milito A, Lugini L, et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One. 2009; 4:e5219.

59. Hoshino A, Costa-Silva B, Shen TL, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015; 527:329–35.
60. Lu Q, Zhang J, Allison R, et al. Identification of extracellular delta-catenin accumulation for prostate cancer detection. Prostate. 2009; 69:411–8.
61. Moon PG, Lee JE, Cho YE, et al. Identification of developmental endothelial locus-1 on circulating extracellular vesicles as a novel biomarker for early breast cancer detection. Clin Cancer Res. 2016; 22:1757–66.

62. Rontogianni S, Synadaki E, Li B, et al. Proteomic profiling of extracellular vesicles allows for human breast cancer subtyping. Commun Biol. 2019; 2:325.

63. Liu Q, Yu Z, Yuan S, et al. Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget. 2017; 8:13048–58.

64. Yuwen DL, Sheng BB, Liu J, Wenyu W, Shu YQ. MiR-146a-5p level in serum exosomes predicts therapeutic effect of cisplatin in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2017; 21:2650–8.
65. Thakur BK, Zhang H, Becker A, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014; 24:766–9.

66. Vagner T, Spinelli C, Minciacchi VR, et al. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J Extracell Vesicles. 2018; 7:1505403.

67. Kahlert C. Liquid biopsy: is there an advantage to analyzing circulating exosomal DNA compared to cfDNA or are they the same? Cancer Res. 2019; 79:2462–5.

68. Lazaro-Ibanez E, Lasser C, Shelke GV, et al. DNA analysis of low- and high-density fractions defines heterogeneous subpopulations of small extracellular vesicles based on their DNA cargo and topology. J Extracell Vesicles. 2019; 8:1656993.
69. Sharma A, Johnson A. Exosome DNA: critical regulator of tumor immunity and a diagnostic biomarker. J Cell Physiol. 2020; 235:1921–32.

70. Mouliere F, Robert B, Arnau Peyrotte E, et al. High fragmentation characterizes tumour-derived circulating DNA. PLoS One. 2011; 6:e23418.

71. Zill OA, Greene C, Sebisanovic D, et al. Cell-free DNA next-generation sequencing in pancreatobiliary carcinomas. Cancer Discov. 2015; 5:1040–8.

72. San Lucas FA, Allenson K, Bernard V, et al. Minimally invasive genomic and transcriptomic profiling of visceral cancers by next-generation sequencing of circulating exosomes. Ann Oncol. 2016; 27:635–41.

73. Song Z, Cai Z, Yan J, Shao YW, Zhang Y. Liquid biopsies using pleural effusion-derived exosomal DNA in advanced lung adenocarcinoma. Transl Lung Cancer Res. 2019; 8:392–400.

74. Lee DH, Yoon H, Park S, et al. Urinary Exosomal and cell-free DNA detects somatic mutation and copy number alteration in urothelial carcinoma of bladder. Sci Rep. 2018; 8:14707.

75. Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011; 11:426–37.

76. Hur JY, Lee JS, Kim IA, Kim HJ, Kim WS, Lee KY. Extracellular vesicle-based EGFR genotyping in bronchoalveolar lavage fluid from treatment-naive non-small cell lung cancer patients. Transl Lung Cancer Res. 2019; 8:1051–60.
77. Bernard V, Kim DU, San Lucas FA, et al. Circulating nucleic acids are associated with outcomes of patients with pancreatic cancer. Gastroenterology. 2019; 156:108–18.

78. Allenson K, Castillo J, San Lucas FA, et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol. 2017; 28:741–7.
79. Karlovich C, Goldman JW, Sun JM, et al. Assessment of EGFR mutation status in matched plasma and tumor tissue of NSCLC patients from a phase I study of rociletinib (CO-1686). Clin Cancer Res. 2016; 22:2386–95.
80. Oxnard GR, Thress KS, Alden RS, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol. 2016; 34:3375–82.

81. Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2016; 2:1014–22.
82. Helman E, Nguyen M, Karlovich CA, et al. Cell-free DNA next-generation sequencing prediction of response and resistance to third-generation EGFR inhibitor. Clin Lung Cancer. 2018; 19:518–30.
83. Brinkman K, Meyer L, Bickel A, et al. Extracellular vesicles from plasma have higher tumour RNA fraction than platelets. J Extracell Vesicles. 2020; 9:1741176.

84. Qi Y, Zha W, Zhang W. Exosomal miR-660-5p promotes tumor growth and metastasis in non-small cell lung cancer. J BUON. 2019; 24:599–607.
85. Zhang Y, Zhang Y, Yin Y, Li S. Detection of circulating exosomal miR-17-5p serves as a novel non-invasive diagnostic marker for non-small cell lung cancer patients. Pathol Res Pract. 2019; 215:152466.

86. Grimolizzi F, Monaco F, Leoni F, et al. Exosomal miR-126 as a circulating biomarker in non-small-cell lung cancer regulating cancer progression. Sci Rep. 2017; 7:15277.

87. Jin X, Chen Y, Chen H, et al. Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin Cancer Res. 2017; 23:5311–9.

88. Hu Z, Chen X, Zhao Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010; 28:1721–6.

89. Krug AK, Enderle D, Karlovich C, et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann Oncol. 2018; 29:700–6.
90. Morgensztern D, Waqar S, Subramanian J, Trinkaus K, Govindan R. Prognostic impact of malignant pleural effusion at presentation in patients with metastatic non-small-cell lung cancer. J Thorac Oncol. 2012; 7:1485–9.

91. William WN Jr, Lin HY, Lee JJ, Lippman SM, Roth JA, Kim ES. Revisiting stage IIIB and IV non-small cell lung cancer: analysis of the surveillance, epidemiology, and end results data. Chest. 2009; 136:701–9.
92. Hooper C, Lee YC, Maskell N; Group BTSPG. Investigation of a unilateral pleural effusion in adults: British Thoracic Society Pleural Disease Guideline 2010. Thorax. 2010; 65 Suppl 2:ii4–17.

93. Lin J, Gu Y, Du R, Deng M, Lu Y, Ding Y. Detection of EGFR mutation in supernatant, cell pellets of pleural effusion and tumor tissues from non-small cell lung cancer patients by high resolution melting analysis and sequencing. Int J Clin Exp Pathol. 2014; 7:8813–22.
94. Shin S, Kim J, Kim Y, Cho SM, Lee KA. Assessment of real-time PCR method for detection of EGFR mutation using both supernatant and cell pellet of malignant pleural effusion samples from non-small-cell lung cancer patients. Clin Chem Lab Med. 2017; 55:1962–9.

95. Qu X, Li Q, Yang J, et al. Double-stranded DNA in exosomes of malignant pleural effusions as a novel DNA source for EGFR mutation detection in lung adenocarcinoma. Front Oncol. 2019; 9:931.

96. Lee JS, Hur JY, Kim IA, et al. Liquid biopsy using the supernatant of a pleural effusion for EGFR genotyping in pulmonary adenocarcinoma patients: a comparison between cell-free DNA and extracellular vesicle-derived DNA. BMC Cancer. 2018; 18:1236.

Go to : 




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download