Abstract
Background
High-risk (HR) human papillomavirus (HPV) is found in a subset of head and neck (HN) squamous cell carcinomas (SCCs). For oropharyngeal SCCs, HR HPV positivity is known to be associated with good prognosis, and a separate staging system for HPV-associated carcinomas using p16 immunohistochemistry (IHC) as a surrogate test has been adopted in the 8th American Joint Committee on Cancer staging system. We examined the HR HPV status and the genotype distribution in five HN subsites.
Methods
Formalin-fixed paraffin-embedded tissue sections were used for p16 IHC and DNA extraction. HPV DNA detection and genotyping were done employing either a DNA chip-based or real-time polymerase chain reaction–based method.
Results
During 2011–2019, a total of 466 SCCs were tested for HPV DNA with 34.1% positivity for HR HPV. Among HN subsites, the oropharynx showed the highest HR HPV prevalence (149/205, 75.1%), followed by the sinonasal tract (3/14, 21.4%), larynx (5/43, 11.6%), hypopharynx (1/38, 2.6%), and oral cavity (1/166, 0.6%). The most common HPV genotype was HPV16 (84.3%) followed by HPV35 (6.9%) and HPV33 (4.4%). Compared with HR HPV status, the sensitivity and specificity of p16 IHC were 98.6% and 94.3% for the oropharynx, and 99.2% and 93.8% for the tonsil, respectively.
Conclusions
Using a Korean dataset, we confirmed that HR HPV is most frequently detected in oropharyngeal SCCs. p16 positivity showed a good concordance with HR HPV DNA for oropharyngeal and especially tonsillar carcinomas. The use of p16 IHC may further be extended to predict HR HPV positivity in sinonasal tract SCCs.
Human papillomavirus (HPV)-associated squamous neoplasia, from simple warts to squamous cell carcinoma (SCC), occurs in various skin and mucosal sites including the uterine cervix, anogenital area, and upper aerodigestive tract of head and neck (HN). Among HPV genotypes, those that can transform the infected cells to malignancy are designated ‘high-risk’ (HR) types, the most well-known of which is HPV16. For the uterine cervix, about 90% of SCCs are associated with HR HPV [1]. In the HN, HR HPV prevalence of SCCs varies among the subsites, the oropharyngeal tumors being predominantly associated with the virus [2,3].
HR HPV infection leads to overexpression of p16 protein as a consequence of viral E7-mediated degradation of retinoblastoma (Rb) protein [4]. Therefore, p16 immunohistochemistry (IHC) in paraffin-embedded tissue has been utilized as a surrogate test for HPV-specific tests, such as detection of HPV DNA and RNA [5]. In the oropharynx, the prognosis of HR HPV-positive SCCs is known to be superior compared with that of HPV-negative tumors [6,7]. p16 positivity alone was also shown to be an independent prognostic factor of oropharyngeal SCCs [5,6], and the recent 8th American Joint Committee on Cancer (AJCC) staging system adopted a separate TNM staging system for ‘HPV-mediated (p16 positive)’ oropharyngeal carcinomas [8].
The incidence of oropharyngeal cancer, most of which are SCCs, has been reported to be increasing worldwide, and this is attributed to a rise in HPV-positive portion of oropharyngeal carcinomas in some studies [3,9-12]. As for the Republic of Korea, a National Health Insurance Service data–based study reported an increase in the incidence of tonsillar cancer during 2002–2015 from 1.1 to 2.4 for men, and from 0.31 to 0.46 for women (per 100,000) [13]. On the other hand, data on HPV-positive fraction of Korean HN SCCs has been published for small cohorts of oropharynx and oral cavity tumors, with a wide range of values [14-24]. Using a recent (2011–2019) and large (n=466) HN SCC dataset and employing HPV DNA polymerase chain reaction (PCR), we aimed to assess the HPV-attributable fraction in each subsite of the oropharynx, oral cavity, larynx, hypopharynx, and sinonasal tract.
The HN SCC dataset of this study were derived from two of Catholic Medical Center hospitals, Seoul St. Mary’s Hospital and Bucheon St. Mary’s Hospital. The pathology file of Seoul St. Mary’s Hospital was searched for HN SCCs that underwent p16 IHC and/or HPV DNA PCR test as ancillary tests during the period from January 2011 to December 2019. A total of 717 HN SCC cases from 717 patients were retrieved, which includes small biopsies as well as resection cases. Bucheon St. Mary’s Hospital dataset consists of 70 HN SCC cases accessioned from January 2011 to December 2019. Thirty-one of Bucheon St. Mary’s Hospital cases had undergone p16 IHC and/or HPV DNA PCR at the time of diagnosis. For the remaining 39 cases for which p16 IHC and/or HPV PCR was not already done, paraffin-embedded tissue blocks were used to perform p16 IHC and HPV PCR. The paraffin blocks of Bucheon St. Mary’s Hospital cases were loaned from Bucheon St. Mary’s Biobank of The Catholic University of Korea (B2019091702). In total, 787 HN SCC cases from the two hospitals were analyzed. Included in the dataset were 28 cases for which p16 IHC and/or HPV PCR were not performed on the primary tumor but on the metastatic tumors of neck lymph nodes (n=23) or on the recurrent tumors (n=5). The p16 and HPV status of the primary tumor were assumed to be the same as in the metastatic/recurrent tumor [25] for these cases. Squamous cell carcinomas of unknown origin, nasopharyngeal carcinomas, and adenosquamous carcinomas were excluded.
Formalin-fixed paraffin-embedded (FFPE) tissue sections of 4-μm thickness were used for IHC employing Ventana BenchMark autostainer (Ventana Medical Systems, Tucson, AZ, USA) in each of the two hospitals. The p16 antibody used during the year 2011–2015 at Seoul St. Mary’s Hospital was a mouse monoclonal antibody (JC8, 1:200, Santa Cruz Biotechnology, Santa Cruz, CA, USA). Cases from year 2016 onward at Seoul St. Mary’s Hospital and all of Bucheon St. Mary’s Hospital were stained with a ready-to-use antibody of E6H4 clone (CINtec Histology, Ventana Medical Systems). p16 IHC result was considered positive when there was diffuse moderate or strong nuclear and cytoplasmic staining in more than 70% of the tumor cells.
DNA was extracted from four 10-μm-thick FFPE tissue sections of each case using QIASymphony DNA mini kit (Qiagen, Hilden, Germany) before 2016, and Maxwell 16 automated system (Promega Corp., Madison, WI, USA) later. HPV DNA detection and genotyping were done using either of the two commercial PCR-based kits. For cases accessioned before March 2018, a DNA chip-based genotyping kit (BMT HPV 9G DNA kit, Biometrix Technology, Chuncheon, Korea) was used. The later cases were tested using PANA RealTyper HPV Kit (PANAGENE Inc., Daejeon, Korea) that employs peptide nucleic acid probe-based multiplex real-time PCR and melting curve analysis. These tests were run at the department of pathology laboratories of Seoul St. Mary’s Hospital, where Bucheon St. Mary’s Hospital specimens were sent out. The HPV genotypes identified by the BMT HPV 9G DNA kit are 14 HR types (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68) and five low-risk (LR) types (6, 11, 34, 40, 42), and the rest of HPV DNA are reported as ‘other’ types. The PANA RealTyper HPV Kit specifies 20 HR types (16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 69, 70, 73, 82) and two LR types (6, 11) while reporting 18 LR types (30, 32, 34, 40, 42, 43, 44, 54, 55, 61, 62, 67, 74, 81, 83, 84, 87, 90) as ‘other’ types. Statistical analyses were done using Excel 2013 (Microsoft Corp., Seattle, WA, USA) and SPSS statistics ver. 20.0 (IBM SPSS, Armonk, NY, USA) programs. A p-value<.05, double-sided, was considered statistically significant.
The dataset of 787 HN SCCs (Table 1) consisted of 271 oral cavity (34.4%), 257 oropharynx (32.7%), 156 larynx (19.8%), 76 hypopharynx (9.7%), and 27 sinonasal tract (3.4%) carcinomas. p16 IHC was done for 771 of 787 cases. HPV DNA PCR was run for 466 cases, including 16 cases devoid of p16 IHC results. p16 positivity was 29.6% (228/771) in HN SCCs being the most prevalent in oropharyngeal carcinomas (184/252, 73.0%) and secondly in sinonasal tract carcinomas (7/25, 28.0%). HR HPV PCR-positivity was detected in 34.1% (159/466) of HN SCCs, 93.7% (149/159) of which was oropharyngeal carcinomas. The HR HPV PCR-positivity among oropharyngeal carcinomas was 72.7%, comparable to its p16-positive rate (73.0%). The prevalence of HR HPV in other subsites were 21.4%, 11.6%, 2.6% and 0.6%, for sinonasal tract, larynx, hypopharynx, and oral cavity, respectively. Among the HR HPV genotypes, HPV16 was overall the most common (84.3%, 134/159). HPV35 and 33 were the second and third most common ones, detected at 6.9% (n=11) and 4.4% (n=7), respectively. HPV58 and HPV18, each, was observed at 1.9% (n=3). HPV18 co-occurred with HPV16 in two of the three cases. Multiple genotypes were detected in seven cases as a combination of HR types (2 of 16 & 18; 16, 51 & 68) or both HR and LR types (16, 52 & 81; 16 & 34; 16, 31, 39 & 6; 58 & 11). HR HPV prevalence in the oropharynx was further stratified according to its four subsites of tonsil (palatine), base of tongue, soft palate, and other oropharynx (posterior wall or unspecified). The tonsil and base of tongue showed high rates of HR HPV positivity (78.9% and 61.3%, respectively) compared with soft palate (0%, 0/5) and other oropharynx (37.5%, 3/8). There were 15 cases (3.2%, 15/466) where only a LR (n=5) or ‘other’ type (n=10) was detected. Cases positive for ‘other’ type were detected with the chip-based genotyping kit, and the HPV type could include either a minority of HR type (26, 53, 69, 70, 73, and 82) or an LR type according to the manufacturer.
The correlation of p16 overexpression with transcriptionallyactive HR HPV infection is well known for oropharyngeal SCCs [5]. We calculated the sensitivity, specificity, positive predictive value (PPV), and negative predictive value of p16 IHC in predicting HR HPV DNA positivity in 450 HN SCCs (Table 2). Differences in these measures were notable among HN subsites. The oropharynx (n=200) demonstrated high rates for the sensitivity, specificity, and PPV: 98.6 and 94.3 and 98.0%, respectively. When limited to the tonsil (n=157), the sensitivity even reached 99.2%, with the specificity of 93.8%. By contrast, the sensitivity was 0% for oral cavity carcinomas (n=159), the only HR HPV-harboring carcinoma being p16-negative. Among the nine p16-positive oral cavity carcinomas, one was positive for LR HPV, and the rest was negative for HPV. The numbers of hypopharyngeal and sinonasal tract carcinomas were not large enough in our dataset to consider their 100% sensitivity values significant.
The incidence of HR HPV-associated oropharyngeal SCCs has been shown to be rising over the past few decades in various parts of the world [9]. Our group had previously published a study on tonsillar SCCs diagnosed during 1994–2010 at Seoul St. Mary’s Hospital [26], in which the proportion of p16-positive cases was 62.0% (n=31 of 50). As p16 IHC is a reliable surrogate test for HR HPV in the oropharynx, we compared the proportion of p16-positive tonsillar SCCs across the time periods 1994–2010 and 2011–2019. The p16-positive fraction for 2011–2019, 80.6% (137 of 170), was significantly different from that of 1994–2010, 62.0% (chi-square test, p=.008), demonstrating an increase in HR HPV-positive tonsillar SCCs cases over a decade of time diagnosed in a tertiary care hospital. However, from 2011 to 2019, a significant increasing trend in the proportion of HR HPV-positive carcinomas was not detected (linear-by-linear association test, p=.097) (Fig. 1). Of note, the proportion of HR HPV-positive tonsillar carcinomas for Bucheon St. Mary’s Hospital cohort of current study was 70.8% (17 of 24), not significantly different from 81.5% (110 of 135) of Seoul St. Mary’s Hospital (chi-square test, p=.269).
Searching the literature for HPV-associated HN SCCs of Korea identified 15 studies, which are summarized in Table 3. Most of these studies dealt with time periods earlier than that of our study, from 1990s to 2000s. Studies of tonsillar SCCs employing HPV DNA PCR reported HR HPV prevalence ranging from 23.4% to 73.5%. A roughly bimodal distribution of previously reported HPV prevalence in tonsillar SCCs could be assumed, one peak at ~30% (n=4) and the other at ~70% (n=3). Those rates at 64.1, 73.1, and 73.5% [17,23,27] were not significantly different from the 78.1% of the current study (chi-square test, p>.05 for each) although derived from cohorts much earlier than (dated 2007 or prior) the current study. On the other hand, rates at 20%–30% level in four studies [18,19,22,28] were far lower compared with the others at 70% level. Another four studies of tonsillar or oropharyngeal SCCs documented p16 IHC results without performing HPV-specific tests; the p16-positive rates were all greater than 50%, from 62.0% to 82.5% [16,20, 26,29]. For other HN subsites, reports of HR HPV prevalence were found for oral cavity and hypopharyngeal carcinomas (Table 3). The oral cavity HR HPV positivity (0.6%) of the current study was the least of all Korean studies while the sample size was the largest (n=166).
The present study is the first to survey the HPV prevalence and p16 positivity for SCCs of multiple HN subsites in Korea. HR HPV was predominantly found in oropharyngeal carcinomas, with 72.7% positivity. Meta-analysis studies have shown overall HPV prevalence of 40%–50% for oropharyngeal carcinomas globally, with the highest rates of 70%–75% belonging to North American and Northern European countries [10,30-32]. It is interesting that the oropharyngeal HPV positivity, 72.3%, of the current Korean dataset corresponds to the upper range of prevalence values observed in some Western populations and is higher than those of other Asian countries (mostly at about 50% or lower) [32,33]. Among oropharyngeal subsites in our series, HR HPV prevalence was highest in the tonsil (78.9%) and second highest in the tongue base (BOT) (59.4%) while zero in the soft palate/uvula. The prevailing association of HR HPV with lymphoepithelial subsites of oropharynx, the tonsil and to a lesser extent the BOT, is well established [33]. p16 positivity demonstrated a good concordance with HR HPV DNA in our series of oropharyngeal carcinomas, especially for tonsillar carcinomas, reassuring the surrogate use of p16 IHC for classification of HPVassociated oropharyngeal carcinomas [8]. As p16 overexpression likely represents transcriptionally-active HPV infection, p16- negative HR HPV DNA positivity may suggest inactive infection or DNA contamination [34,35]. Such incidences were rare for oropharyngeal carcinomas in our series (1.6%, 2 out of 140 HR HPV-positive cases).
HR HPV has been recognized as a carcinogenic agent and also a favorable prognostic factor for oropharyngeal carcinomas, but the significance of HPV in non-oropharyngeal HN SCCs are controversial [7,36]. We observed the second highest HR HPV prevalence (21.4%) in sinonasal tract SCCs with a good concordance (100%) between p16 and HR HPV in a series of 12 cases. This corroborates the previous demonstration in sinonasal SCCs of 20%–30% HR HPV prevalence with reliable p16 IHC for HR HPV [37-39]. For oral cavity and laryngeal SCCs, the concordance between p16 and HR HPV status was poor in the current study, which is also in line with previous findings [41]. We observed the lowest HR HPV positivity in oral cavity SCCs, at 0.6% out of 166 cases tested. This is lower than the rates reported in other Korean studies (Table 3) and the prevalence summarized for oral cavity carcinomas in meta-analysis studies (24%–58%) [42]. As we obtained a high rate of HR HPV positivity for oropharyngeal carcinomas backed by a good concordance with p16 positivity, the low rate in oral cavity carcinomas is not likely due to a low sensitivity of the HPV detection methods we used. One possibility for higher HR HPV rates of oral cavity carcinomas in other studies could be inadvertent classification of HR HPVpositive base of tongue cancers as oral cavity (mobile tongue) cancers. Additional larger cohort studies are awaited to evaluate the proportion of HR HPV-positive oral cavity SCCs in Korea.
We noted that the proportion of HPV-associated tonsillar SCCs significantly increased, from 62.0% to 79.5%, over a decade of time by comparing the p16-positive proportion of the previous and current case series from a single hospital. The seven earlier Korean studies documenting HPV DNA status in tonsillar SCCs showed either a rate comparable to the 78.9% of our series or a much lower rate at 20%–30% (Table 3). If not attributable to a lower sensitivity of the particular HPV detection method used in the latter group of studies, the explanation for the lower HPV prevalence may be sought in the cohort characteristics. HPV-positive oropharyngeal SCCs are known to be associated with certain epidemiologic factors, such as younger age at onset, higher socioeconomic status, less tobacco/alcohol consumption and a greater number of lifetime sexual partners in Western studies [43]. Two of the four studies reporting lower HPV prevalence provided p16 positivity rates that did not vary greatly from their HPV detection rates [19,28]; this supports the possibility of true heterogeneity in HPV prevalence among different Korean cohorts. Further studies using a large cohort with consideration of socioeconomic risk factors are warranted to examine the prevalence of HPV-associated tonsillar/oropharyngeal carcinomas in Korea.
HPV16 was the most common HR HPV genotype both among non-oropharyngeal HN SCCs (90%, 9/10) and among oropharyngeal SCCs (83.9%) (Table 1) in our dataset as it has been known [2,31]. For the oropharyngeal SCC series, the second most common HR HPV type was HPV35 (7.4%, 11/149). Literature on HPV types of oropharyngeal SCCs in other countries shows that HPV16 almost always comprised 85%–90% and that HPV33 and HPV35 frequently ranked second or third comprising usually less than 5% each [11,44-46]. In the six Korean tonsillar SCC studies reporting HPV genotypes (Table 3), HPV33 and/or HPV18 were often second most frequent to HPV16. Of note is that the recently developed 9-valent HPV vaccine targets seven HR types including HPV16, 18, 31, 33, 45, 52, and 58, thus not likely offering protection against HPV35. More studies on HPV genotypes in Korean oropharyngeal carcinomas are anticipated to evaluate the prevalence of HPV35.
In conclusion, we have examined the prevalence of HR HPV in 466 HN SCCs according to the five subsites of oral cavity, oropharynx, larynx, hypopharynx and sinonasal tract. The oropharyngeal SCCs showed the highest rate of HR HPV positivity among the HN subsites at 72.3%, and the rate peaked at 78.9% when only tonsillar carcinomas were considered. The proportion of HR HPV-positive tonsillar carcinomas significantly increased over a decade of time in a single hospital. As it has been known, p16 immunopositivity showed a good concordance with HR HPV DNA for oropharyngeal and especially tonsillar carcinomas. The use of p16 IHC may further be extended to predict HR HPV positivity in sinonasal tract SCCs.
Notes
Ethics Statement
The study protocol was approved by the Institutional Review Board of the Catholic Medical Center (XC19SEDI0066), with a waiver of informed consent.
Author contributions
Conceptualization: YSL, YK. Data curation: YHJ, MSK, YSL, YK. Formal Analysis: YK. Investigation: YSL, YK. Methodology: YSL, YK. Project administration: YSL, YK. Resources: YHJ, MSK, YSL, YK. Software: YK. Validation: YSL, YK. Visualization: YK. Funding acquisition: YK. Writing—original draft: YK. Writing—review & editing: YK, YSL, YHJ, MSK.
References
1. de Sanjose S, Quint WG, Alemany L, et al. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol. 2010; 11:1048–56.
2. de Martel C, Plummer M, Vignat J, Franceschi S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int J Cancer. 2017; 141:664–70.

3. Gillison ML, Chaturvedi AK, Anderson WF, Fakhry C. Epidemiology of human papillomavirus-positive head and neck squamous cell carcinoma. J Clin Oncol. 2015; 33:3235–42.

4. Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010; 10:550–60.

5. Lewis JS Jr. p16 immunohistochemistry as a standalone test for risk stratification in oropharyngeal squamous cell carcinoma. Head Neck Pathol. 2012; 6 Suppl 1:S75–82.

6. Rischin D, Young RJ, Fisher R, et al. Prognostic significance of p16INK4A and human papillomavirus in patients with oropharyngeal cancer treated on TROG 02.02 phase III trial. J Clin Oncol. 2010; 28:4142–8.
7. Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010; 363:24–35.

8. Amin MB, Edge SB, Greene FL, et al. AJCC cancer staging manual. 8th ed. New York: Springer;2017.
9. Castellsague X, Alemany L, Quer M, et al. HPV involvement in head and neck cancers: comprehensive assessment of biomarkers in 3680 patients. J Natl Cancer Inst. 2016; 108:djv403.
10. Mehanna H, Beech T, Nicholson T, et al. Prevalence of human papillomavirus in oropharyngeal and nonoropharyngeal head and neck cancer: systematic review and meta-analysis of trends by time and region. Head Neck. 2013; 35:747–55.
11. Chaturvedi AK, Engels EA, Pfeiffer RM, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011; 29:4294–301.

12. Chaturvedi AK, Anderson WF, Lortet-Tieulent J, et al. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol. 2013; 31:4550–9.

13. Oh JK, Choi HY, Han M, Lee JK, Min KJ, Ki M. Prevalence of human papillomavirus-related diseases in the Republic of Korea: a cross-sectional study. Sex Transm Infect. 2019; 95:292–9.

14. Joo YH, Jung CK, Sun DI, Park JO, Cho KJ, Kim MS. High-risk human papillomavirus and cervical lymph node metastasis in patients with oropharyngeal cancer. Head Neck. 2012; 34:10–4.

15. Kim HS, Seo MH, Kim SM, et al. Prevalence of human papillomavirus infection in the Korean oral cancer patients. J Korean Assoc Maxillofac Plast Reconstr Surg. 2013; 35:227–35.

16. Kim HS, Lee JY, Lim SH, et al. Association between PD-L1 and HPV status and the prognostic value of PD-L1 in oropharyngeal squamous cell carcinoma. Cancer Res Treat. 2016; 48:527–36.

17. Kim SH, Koo BS, Kang S, et al. HPV integration begins in the tonsillar crypt and leads to the alteration of p16, EGFR and c-myc during tumor formation. Int J Cancer. 2007; 120:1418–25.

18. Kim Y, Jeong EH, Min BW, et al. HPV genotyping in squamous cell carcinoma of upper aerodigestive tract. Korean J Pathol. 2010; 44:483–7.

19. Kwon MJ, Kim DH, Park HR, et al. Frequent hepatocyte growth factor overexpression and low frequency of c-Met gene amplification in human papillomavirus-negative tonsillar squamous cell carcinoma and their prognostic significances. Hum Pathol. 2014; 45:1327–38.

20. Lee M, Kim SB, Lee SW, et al. Human papillomavirus prevalence and cell cycle related protein expression in tonsillar squamous cell carcinomas of Korean patients with clinicopathologic analysis. Korean J Pathol. 2013; 47:148–57.

21. Lee SY, Cho NH, Choi EC, et al. Relevance of human papilloma virus (HPV) infection to carcinogenesis of oral tongue cancer. Int J Oral Maxillofac Surg. 2010; 39:678–83.

22. No JH, Sung MW, Hah JH, et al. Prevalence and prognostic value of human papillomavirus genotypes in tonsillar squamous cell carcinoma: a Korean multicenter study. Cancer. 2015; 121:535–44.

23. Park WS, Ryu J, Cho KH, et al. Human papillomavirus in oropharyngeal squamous cell carcinomas in Korea: use of G1 cycle markers as new prognosticators. Head Neck. 2012; 34:1408–17.

24. Shin KH, Park KH, Hong HJ, et al. Prevalence of microsatellite instability, inactivation of mismatch repair genes, p53 mutation, and human papillomavirus infection in Korean oral cancer patients. Int J Oncol. 2002; 21:297–302.

25. Vainshtein J, McHugh JB, Spector ME, et al. Human papillomavirus-related oropharyngeal cancer: HPV and p16 status in the recurrent versus parent tumor. Head Neck. 2015; 37:8–11.

26. Song JS, Kim MS, Park JW, Lee YS, Kang CS. Expression of human papillomavirus-related proteins and its clinical implication in tonsillar squamous cell carcinoma. Korean J Pathol. 2012; 46:177–86.

27. Oh TJ, Kim CJ, Woo SK, et al. Development and clinical evaluation of a highly sensitive DNA microarray for detection and genotyping of human papillomaviruses. J Clin Microbiol. 2004; 42:3272–80.

28. Kim MJ, Ki MS, Kim K, et al. Different protein expression associated with chemotherapy response in oropharyngeal cancer according to HPV status. BMC Cancer. 2014; 14:824.

29. Lee J, Chang JS, Kwon HJ, Kim SH, Shin SJ, Keum KC. Impact of p16 expression in oropharyngeal cancer in the postoperative setting: the necessity of re-evaluating traditional risk stratification. Jpn J Clin Oncol. 2016; 46:911–8.

30. Abogunrin S, Di Tanna GL, Keeping S, Carroll S, Iheanacho I. Prevalence of human papillomavirus in head and neck cancers in European populations: a meta-analysis. BMC Cancer. 2014; 14:968.

31. Ndiaye C, Mena M, Alemany L, et al. HPV DNA, E6/E7 mRNA, and p16INK4a detection in head and neck cancers: a systematic review and meta-analysis. Lancet Oncol. 2014; 15:1319–31.

32. Shaikh MH, McMillan NA, Johnson NW. HPV-associated head and neck cancers in the Asia Pacific: a critical literature review and meta-analysis. Cancer Epidemiol. 2015; 39:923–38.
33. Haeggblom L, Ramqvist T, Tommasino M, Dalianis T, Nasman A. Time to change perspectives on HPV in oropharyngeal cancer: a systematic review of HPV prevalence per oropharyngeal sub-site the last 3 years. Papillomavirus Res. 2017; 4:1–11.

34. Prigge ES, Arbyn M, von Knebel Doeberitz M, Reuschenbach M. Diagnostic accuracy of p16(INK4a) immunohistochemistry in oropharyngeal squamous cell carcinomas: a systematic review and meta-analysis. Int J Cancer. 2017; 140:1186–98.
35. Qureishi A, Mawby T, Fraser L, Shah KA, Moller H, Winter S. Current and future techniques for human papilloma virus (HPV) testing in oropharyngeal squamous cell carcinoma. Eur Arch Otorhinolaryngol. 2017; 274:2675–83.

36. International Agency for Research on Cancer. Biological agents. IARC monographs on the evaluation of carcinogenic risks to humans. Vol. 100B. Lyon: IARC Press;2012. p. 275–6.
37. Bishop JA, Guo TW, Smith DF, et al. Human papillomavirus-related carcinomas of the sinonasal tract. Am J Surg Pathol. 2013; 37:185–92.

38. Laco J, Sieglova K, Vosmikova H, et al. The presence of high-risk human papillomavirus (HPV) E6/E7 mRNA transcripts in a subset of sinonasal carcinomas is evidence of involvement of HPV in its etiopathogenesis. Virchows Arch. 2015; 467:405–15.

39. Larque AB, Hakim S, Ordi J, et al. High-risk human papillomavirus is transcriptionally active in a subset of sinonasal squamous cell carcinomas. Mod Pathol. 2014; 27:343–51.

40. Joo YH, Lee YS, Cho KJ, et al. Characteristics and prognostic implications of high-risk HPV-associated hypopharyngeal cancers. PLoS One. 2013; 8:e78718.

41. Lewis JS Jr, Ukpo OC, Ma XJ, et al. Transcriptionally-active highrisk human papillomavirus is rare in oral cavity and laryngeal/hypopharyngeal squamous cell carcinomas: a tissue microarray study utilizing E6/E7 mRNA in situ hybridization. Histopathology. 2012; 60:982–91.
42. Syrjanen S, Syrjanen K. HPV in head and neck carcinomas: different HPV profiles in oropharyngeal carcinomas: why? Acta Cytol. 2019; 63:124–42.
43. Benard VB, Johnson CJ, Thompson TD, et al. Examining the association between socioeconomic status and potential human papillomavirus-associated cancers. Cancer. 2008; 113(10 Suppl):2910–8.

44. Goodman MT, Saraiya M, Thompson TD, et al. Human papillomavirus genotype and oropharynx cancer survival in the United States of America. Eur J Cancer. 2015; 51:2759–67.

Fig. 1.
High-risk (HR) human papillomavirus (HPV) prevalence and p16 positivity in tonsillar squamous cell carcinomas (SCCs) across years at diagnosis. Trends in the proportion of HR HPV-positive (black line) and p16-positive (grey line) newly diagnosed tonsillar SCCs for the Seoul St. Mary’s Hospital cohort. Numbers above or below a datapoint indicate the total number of tests done in the corresponding year (black, HPV polymerase chain reaction; grey, p16 immunohistochemistry).
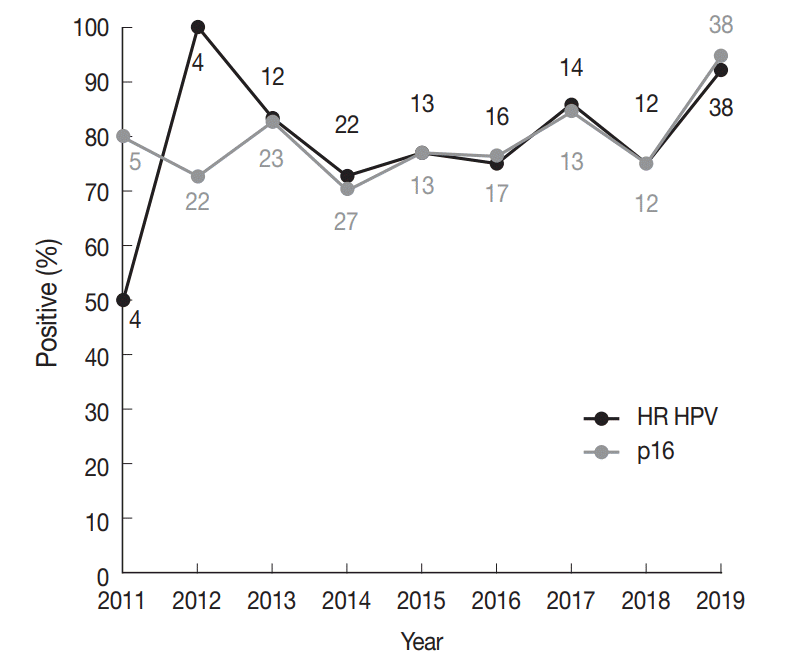
Table 1.
Frequency of p16-positive and HR HPV-positive cases in head and neck subsites
| Subsite | No. | p16 IHCa | p16+ | HPV DNA PCRb | HPV+c | HR HPV+ | HR genotype (n) | |
|---|---|---|---|---|---|---|---|---|
| Oral cavity | 271 | 264 | 20 (7.6) | 166 | 5 (3.0) | 1 (0.6) | - | |
| Tongue | 185 | 180 | 15 (8.3) | 115 | 2 (1.7) | 0 | - | |
| Mouth floor | 20 | 20 | 2 (10.0) | 14 | 2 (14.3) | 1 (7.1) | 16 & 31 & 39 (1) | |
| Retromolar trigone | 16 | 15 | 1 (6.7) | 10 | 0 | 0 | - | |
| Gingiva | 17 | 17 | 0 | 6 | 0 | 0 | - | |
| Buccal mucosa | 25 | 24 | 2 (8.3) | 16 | 1 (6.3) | 0 | - | |
| Hard palate | 7 | 7 | 0 | 4 | 0 | 0 | - | |
| Mucosal lip | 1 | 1 | 0 | 1 | 0 | 0 | - | |
| Oropharynx | 257 | 252 | 184 (73.0) | 205 | 154 (75.1) | 149 (72.7) | - | |
| Tonsil | 206 | 202 | 157 (77.7) | 161 | 130 (80.7) | 127 (78.9) | 16 (105), 16 & 18 (1), 16 & 52 (1), 18 (1), 33 (6), 35 (10), 58 (3), | |
| Tongue base | 36 | 35 | 22 (62.9) | 31 | 19 (61.3) | 19 (61.3) | 16 (16), 33 (1), 58 (1), 69 (1) | |
| Soft palate and uvula | 6 | 6 | 1 (16.7) | 5 | 1 (20.0) | 0 | - | |
| Other oropharynx | 9 | 9 | 4 (44.4) | 8 | 4 (50.0) | 3 (37.5) | 16 (2), 35 (1) | |
| Sinonasal tractd | 27 | 25 | 7 (28.0) | 14 | 5 (35.7) | 3 (21.4) | 16 (2), 69 (1) | |
| Larynx | 156 | 154 | 11 (7.1) | 43 | 8 (18.6) | 5 (11.6) | 16 (3), 16 & 51 & 68 (1), 16 & 18 (1) | |
| Hypopharynx | 76 | 75 | 6 (8.0) | 38 | 2 (5.3) | 1 (2.6) | 16 (1) | |
| Total | 787 | 771 | 228 (29.6) | 466 | 174 (37.3) | 159 (34.1) | 16 (134), 35 (11), 33 (7), 18 (3), 58 (4) | |
Table 2.
Concordance between p16 IHC results and HR HPV status
| No. | p16+ | HR HPV+ | p16+ and HR HPV+ | p16- and HR HPV+ | p16+ and HR HPV– | Sensitivitya (%) | Specificitya (%) | PPVa (%) | NPVa (%) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Oral cavity | 159 | 9 | 1 | 0 | 1 | 9 | 0 | 94.3 | 0 | 99.3 | |
| Oropharynx | 200 | 148 | 147 | 145 | 2b | 3 | 98.6 | 94.3 | 98.0 | 96.2 | |
| Tonsil | 157 | 126 | 125 | 124 | 1 | 2 | 99.2 | 93.8 | 98.4 | 96.8 | |
| BOT | 30 | 19 | 19 | 18 | 1 | 1 | 94.7 | 90.9 | 94.7 | 90.9 | |
| Larynx | 42 | 4 | 5 | 2 | 3 | 2 | 40.0 | 94.6 | 50.0 | 92.1 | |
| Hypopharynx | 37 | 1 | 1 | 1 | 0 | 0 | 100 | 100 | 100 | 100 | |
| Sinonasal tract | 12 | 3 | 3 | 3 | 0 | 0 | 100 | 100 | 100 | 100 | |
| Overall | 450 | 165 | 157 | 151 | 6 | 14 | 96.2 | 95.2 | 91.5 | 97.9 | |
Table 3.
HR HPV prevalence of head and neck squamous cell carcinomas in Korean studies
| Study | Site | Sampling perioda | HPV detection method | HR HPV prevalence, n (%) | HPV genotype (%) |
|---|---|---|---|---|---|
| Oh et al. (2004) [27] | Tonsil | NS | PCRb | 25/39 (64.1) | 16 (92), 33 (4), 58 (4) |
| Kim et al. (2007) [17] | Tonsil | 1995–2005 | PCR | 38/52 (73.1) | 16 (89.5), 18 (2.6), 33 (2.6), 35 (2.6), 58 (2.6) |
| Kim et al. (2010) [18] | Tonsil | 1999–2004 | PCR | 12/47 (25.5) | 16 (100), 18 (8.3)d |
| Park et al. (2012) [23] | OP (tonsil, BOT, SP, P) | 2002–2007 | PCR, p16c | OP (PCR): 53/93 (57.0) | 16 (95.6), 18 (2.2), 33 (2.2) |
| OP (p16): 46/93 (49.5) | |||||
| Tonsil (PCR): 36/49 (73.5) | |||||
| Song et al. (2012) [26] | Tonsil | 1994–2010 | ISH, p16 | ISH: 20/52 (38.5) | NA |
| p16: 31/50 (62.0) | |||||
| Lee et al. (2013) [20] | Tonsil | 2000–2010 | ISH, p16 | ISH: 59/89 (66.3) | NA |
| p16: 74/89 (83.1) | |||||
| Kwon et al. (2014) [19] | Tonsil | 1997–2010 | PCR, p16 | PCR: 28/79 (35.4) | 16 (100) |
| p16: 31/79 (39.2) | |||||
| No et al. (2015) [22] | Tonsil | 1998–2009 | PCR | 41/175 (23.4) | 16 (43.9), 18 (43.9) |
| Kim et al. (2014) [28] | OP (tonsil, BOT, SP) | 2004–2011 | PCR, p16 | OP (PCR): 21/74 (28.4) | 16 (76.2), 18 (28.6)d, 33 (4.7), 35 (4.7) |
| OP (p16): 35/74 (47.3) | |||||
| Tonsil (PCR): 15/47 (31.9) | |||||
| Tonsil (p16): 24/47 (51.1) | |||||
| Lee et al. (2016) [29] | OP (tonsil, BOT, SP) | 2001–2011 | p16 | 104/126 (82.5)e | NA |
| Kim et al. (2016) [16] | OP (tonsil, BOT, SP, P) | 2002–2013 | p16 | 89/133 (66.9)e | NA |
| Current study | OP (tonsil, BOT, SP, P) | 2011–2019 | PCR, p16 | OP (PCR): 149/205 (72.7) | 16 (83.9), 35 (7.4), 33 (4.7), 58 (2.7), 18 (1.3), 52 (0.7) |
| OP (p16): 184/252 (73.0) | |||||
| Tonsil (PCR): 127/161 (78.9) | |||||
| Tonsil (p16): 147/189 (77.8) | |||||
| Joo et al. (2013) [40] | Hypopharynx | 1996–2011 | ISH | 7/64 (10.9) | NA |
| Shin et al. (2002) [24] | Oral cavity | NS | HPV16, 18, 33-specific PCR | 11/76 (14.5) | 16 (36.4), 18 (72.7), 33 (18.2) |
| Lee et al. (2010) [21] | Oral cavity | 1995–2005 | PCR | 12/36 (33.3) | 16 (91.7), |
| Kim et al. (2010) [18] | Oral cavity | 1999–2004 | PCR | 1/22 (4.5) | 16 (100) |
| Joo et al. (2012) [14] | Oral cavity | 1994–2009 | ISH | 6/90 (6.7) | NA |
| Kim et al. (2013) [15] | Oral cavity | 2010–2012 | PCR | 2/86 (2.3) | 16 (100) |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download