MATERIALS AND METHODS
Characterization of patients and sample acquisition
Next-generation sequencing
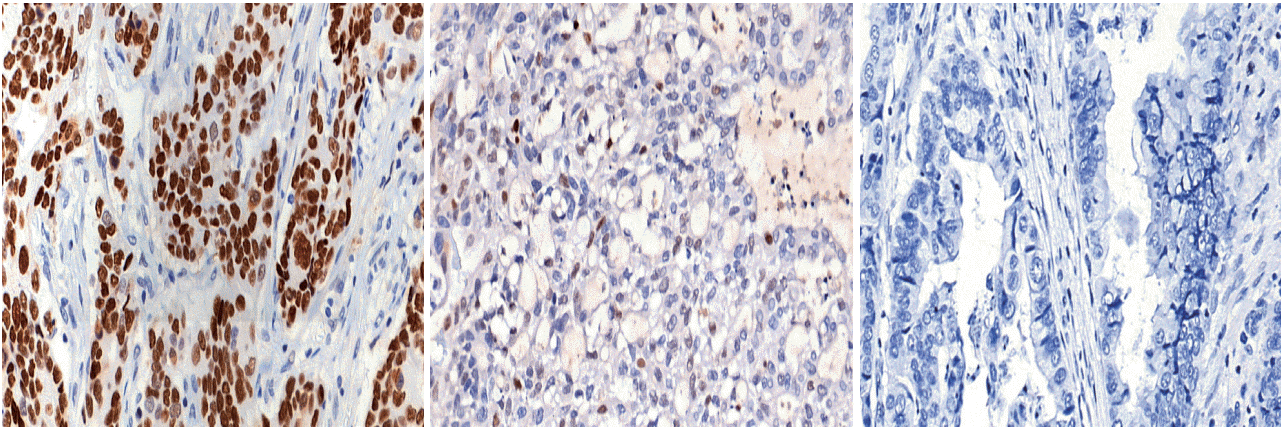
IHC staining
EBV in-situ hybridization
Microsatellite instability analysis
Statistical analyses
RESULTS
Gene mutation and protein expression correlation
Table 1.
| TP53 mutation |
p53 expression by IHC |
Total | p-value | ||
|---|---|---|---|---|---|
| Strong | Negative | Weak | |||
| Mutation status | < .001 | ||||
| Wild-type | 1 (2.9) | 12 (44.4) | 55 (93.2) | 68 (56.7) | |
| Mutation present | 33 (97.1) | 15 (55.6) | 4 (6.8) | 52 (43.3) | |
| Variant summary | < .001 | ||||
| Wild-type | 1 (2.9) | 12 (44.4) | 55 (93.2) | 68 (56.7) | |
| Missense | 30 (88.2) | 0 | 3 (5.1) | 33 (27.5) | |
| Other | 3 (8.9) | 15 (55.6) | 1 (1.7) | 19 (15.8) | |
| Stop-gained | 2 (5.9) | 3 (11.1) | 1 (1.7) | 6 (5.0) | |
| Splice region | 0 | 5 (18.5) | 0 | 5 (4.2) | |
| Frameshift | 0 | 7 (25.9) | 0 | 7 (5.8) | |
| In-frame deletion | 1 (2.9) | 0 | 0 | 1 (0.8) | |
| Clinical significancea | < .001 | ||||
| Wild-type | 1 (2.9) | 12 (44.4) | 55 (93.2) | 68 (56.7) | |
| Pathogenic or likely pathogenic | 22 (64.7) | 13 (48.1) | 2 (3.4) | 37 (30.1) | |
| Uncertain significance | 5 (14.7) | 2 (7.4) | 1 (1.7) | 8 (6.7) | |
| Conflicting interpretation | 6 (17.6) | 0 | 1 (1.7) | 7 (5.8) | |
| Total | 34 | 27 | 59 | 120 | |
Table 2.
| Case No. | Effect | Nucleic acid alteration | Amino acid alteration | Clinical significancea |
|---|---|---|---|---|
| 1 | Missense_variant | c.422G > A | p.Cys141Tyr | Pathogenic or likely pathogenic |
| 2 | Missense_variant | c.422G > T | p.Cys141Phe | Pathogenic or likely pathogenic |
| 3 | Missense_variant | c.455C > T | p.Pro152Leu | Pathogenic or likely pathogenic |
| 4 | Missense_variant | c.524G > A | p.Arg175His | Pathogenic or likely pathogenic |
| 5 | Missense_variant | c.535C > G | p.His179Asp | Pathogenic or likely pathogenic |
| 6 | Missense_variant | c.542G > A | p.Arg181His | Pathogenic or likely pathogenic |
| 7 | Missense_variant | c.659A > G | p.Tyr220Cys | Pathogenic or likely pathogenic |
| 8 | Missense_variant | c.659A > G | p.Tyr220Cys | Pathogenic or likely pathogenic |
| 9 | Missense_variant | c.701A > G | p.Tyr234Cys | Pathogenic or likely pathogenic |
| 10 | Missense_variant | c.725G > A | p.Cys242Tyr | Pathogenic or likely pathogenic |
| 11 | Missense_variant | c.734G > A | p.Gly245Asp | Pathogenic or likely pathogenic |
| 12 | Missense_variant | c.742C > T | p.Arg248Trp | Pathogenic or likely pathogenic |
| 13 | Missense_variant | c.742C > T | p.Arg248Trp | Pathogenic or likely pathogenic |
| 14 | Missense_variant | c.743G > A | p.Arg248Gln | Pathogenic or likely pathogenic |
| 15 | Missense_variant | c.772G > A | p.Glu258Lys | Pathogenic or likely pathogenic |
| 16 | Missense_variant | c.817C > T | p.Arg273Cys | Pathogenic or likely pathogenic |
| 17 | Missense_variant | c.817C > T | p.Arg273Cys | Pathogenic or likely pathogenic |
| 18 | Missense_variant | c.818G > A | p.Arg273His | Pathogenic or likely pathogenic |
| 19 | Missense_variant | c.818G > A | p.Arg273His | Pathogenic or likely pathogenic |
| 20 | Missense_variant | c.818G > A | p.Arg273His | Pathogenic or likely pathogenic |
| 21 | Missense_variant | c.380C > T | p.Ser127Phe | Conflicting interpretations of pathogenicity |
| 22 | Missense_variant | c.473G > C | p.Arg158Pro | Conflicting interpretations of pathogenicity |
| 23 | Missense_variant | c.481G > A | p.Ala161Thr | Conflicting interpretations of pathogenicity |
| 24 | Missense_variant | c.613T > C | p.Tyr205His | Conflicting interpretations of pathogenicity |
| 25 | Missense_variant | c.796G > A | p.Gly266Arg | Conflicting interpretations of pathogenicity |
| 26 | Missense_variant | c.796G > A | p.Gly266Arg | Conflicting interpretations of pathogenicity |
| 27 | Missense_variant | c.1015G > A | p.Glu339Lys | Conflicting interpretations of pathogenicity |
| 28 | Missense_variant | c.329G > A | p.Arg110His | Uncertain significance |
| 29 | Missense_variant | c.380C > A | p.Ser127Tyr | Uncertain significance |
| 30 | Missense_variant | c.476C > T | p.Ala159Val | Uncertain significance |
| 31 | Missense_variant | c.797G > T | p.Gly266Val | Uncertain significance |
| 32 | Missense_variant | c.400T > G | p.Phe134Val | Uncertain significance |
| 33 | Missense_variant | c.470T > G | p.Val157Gly | Uncertain significance |
| 34 | Frameshift_variant | c.331_332insAG | p.Leu111fs | Pathogenic or likely pathogenic |
| 35 | Frameshift_variant | c.381_391delCCCTGCCCTCA | p.Pro128fs | Pathogenic or likely pathogenic |
| 36 | Frameshift_variant | c.635_669delTTCGACATAGTGTGGTG GTGCCCTATGAGCCGCCT | p.Phe212fs | Pathogenic or likely pathogenic |
| 37 | Frameshift_variant | c.660_661delTG | p.Tyr220fs | Pathogenic or likely pathogenic |
| 38 | Frameshift_variant | c.747delG | p.Arg249fs | Pathogenic or likely pathogenic |
| 39 | Frameshift_variant | c.1169delC | p.Pro390fs | Pathogenic or likely pathogenic |
| 40 | Frameshift_variant | c.778_779delTC | p.Ser260fs | Uncertain significance |
| 41 | Conservative_inframe_deletion | c.529_546delCCCCACCATGAGCGCTGC | p.Pro177_Cys182del | Pathogenic or likely pathogenic |
| 42 | Stop_gained | c.159G > A | p.Trp53* | Pathogenic or likely pathogenic |
| 43 | Stop_gained | c.437G > A | p.Trp146* | Pathogenic or likely pathogenic |
| 44 | Stop_gained | c.586C > T | p.Arg196* | Pathogenic or likely pathogenic |
| 45 | Stop_gained | c.637C > T | p.Arg213* | Pathogenic or likely pathogenic |
| 46 | Stop_gained | c.1024C > T | p.Arg342* | Pathogenic or likely pathogenic |
| 47 | Stop_gained | c.1024C > T | p.Arg342* | Pathogenic or likely pathogenic |
| 48 | Splice_region_variant&synonymous_variant | c.375G > A | p.Thr125Thr | Pathogenic or likely pathogenic |
| 49 | Splice_region_variant&synonymous_variant | c.375G > A | p.Thr125Thr | Pathogenic or likely pathogenic |
| 50 | Splice_region_variant&synonymous_variant | c.375G > C | p.Thr125Thr | Pathogenic or likely pathogenic |
| 51 | Splice_acceptor_variant&intron_variant | c.920 - 1G > A | Pathogenic or likely pathogenic | |
| 52 | Splice_donor_variant&intron_variant | c.96 + 1G > A | Uncertain significance (no report) |
Sensitivity, specificity, and accuracy of p53 IHC for predicting TP53 mutations
Table 3.
Clinicopathological variables and protein expression correlations
Table 4.
Values are presented as number (%).
NS, nonsynonymous; Other, other type mutation; wild, wild-type; WHO, World Health Organization; WD, well-differentiated; MD, moderately differentiated; PD, poorly differentiated; PCC, poorly cohesive carcinoma; EBV, Epstein-Barr virus; MSI, microsatellite instability; MSS, microsatellite stable; MSI-L, microsatellite instability-low; MSI-H, microsatellite instability-high.
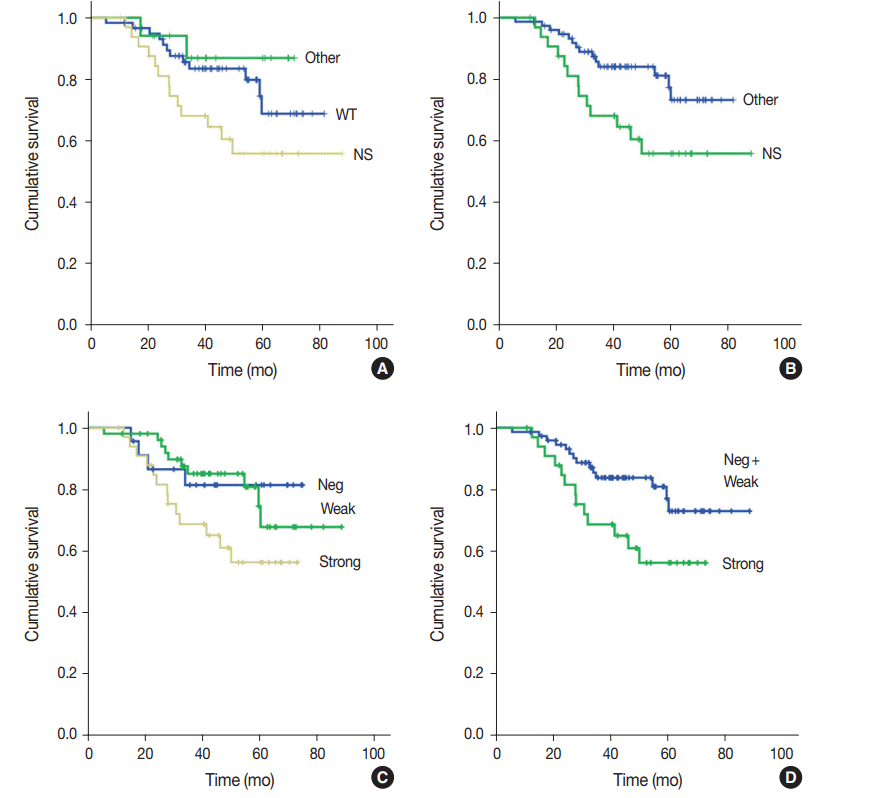
Survival analysis
Fig. 2.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download