This article has been
cited by other articles in ScienceCentral.
Abstract
Background
Microsatellite instability (MSI) analysis is becoming increasingly important in many types of tumor including colorectal cancer (CRC). The commonly used MSI tests are either time-consuming or labor-intensive. A fully automated MSI test, the Idylla MSI assay, has recently been introduced. However, its diagnostic performance has not been extensively validated in clinical CRC samples.
Methods
We evaluated 133 samples whose MSI status had been rigorously validated by standard polymerase chain reaction (PCR), clinical next-generation sequencing (NGS) cancer panel test, or both. We evaluated the diagnostic performance of the Idylla MSI assay in terms of sensitivity, specificity, and positive and negative predictive values, as well as various sample requirements, such as minimum tumor purity and the quality of paraffin blocks.
Results
Compared with the gold standard results confirmed through both PCR MSI test and NGS, the Idylla MSI assay showed 99.05% accuracy (104/105), 100% sensitivity (11/11), 98.94% specificity (93/94), 91.67% positive predictive value (11/12), and 100% negative predictive value (93/93). In addition, the Idylla MSI assay did not require macro-dissection in most samples and reliably detected MSI-high in samples with approximately 10% tumor purity. The total turnaround time was about 150 minutes and the hands-on time was less than 2 minutes.
Conclusions
The Idylla MSI assay shows good diagnostic performance that is sufficient for its implementation in the clinic to determine the MSI status of at least the CRC samples. In addition, the fully automated procedure requires only a few slices of formalin-fixed paraffin-embedded tissue and might greatly save time and labor.
Keywords: Microsatellite instability, Polymerase chain reaction, Idylla MSI test, Diagnostic performance, Tumor purity
Microsatellites, also known as short tandem repeats, are repetitive sequences usually composed of one to six base pairs, repeating from 5 to 50 times and accounting for about 3% of the whole human genome. Due to this repeated structure, DNA sequence mismatches are likely to occur during DNA replication. Mismatch repair (MMR) is a DNA repair system that can correct the errors. Failure to correct these errors during DNA replication results in a variation in the length of the repeat region, which is called microsatellite instability (MSI) [
1]. MSI has been identified in about 15%–20% of colorectal cancer (CRC) which may be sporadic (12%–14%) or Lynch syndrome-associated (1%–3%) [
2]. In sporadic colorectal cases, the tumors are mainly caused by epigenetic silencing of the
MLH1 gene [
3] promoter by acquired hypermethylation, which is in association with a high CpG island methylation phenotype [
4]. It accounts for approximately 70%–90% of CRC with MSI [
1,
5]. In Lynch syndrome, the tumors with MSI are frequently caused by autosomal dominant germline mutations in the MMR genes [
1].
Currently, it is becoming increasingly important to determine the MSI status in patients with CRC because MSI–high (MSI-H) CRC is often associated with Lynch syndrome and MMR deficient CRCs can clinically benefit from immune checkpoint inhibitors [
6]. The MSI status can be detected by immunohistochemistry (IHC) for MMR proteins or polymerase chain reaction (PCR)–based MSI tests, which are currently considered the gold standard method. However, this method requires normal tissues, and it is time-consuming and labor-intensive. Recently, next-generation sequencing (NGS) has been reported to successfully detect the MSI status [
7,
8]. This can be done by either direct sequencing of the microsatellite loci or analysis of the total mutation burden or proportion of insertion/deletion mutations. However, NGS experiments are expensive and only available in specialized laboratories.
In this study, we tested Idylla MSI assay in terms of its diagnostic performance, the minimum requirements for tumor purity, and turnaround time through the examination of the surgically resected samples or colonoscopic biopsy CRC samples, the MSI status of which were rigorously confirmed through either standard PCR fragment analysis or clinical NGS test.
MATERIALS AND METHODS
Sample selection
Formalin-fixed paraffin-embedded (FFPE) tumor tissues from 115 pathologically confirmed CRC patients who underwent surgical resection (n=109) or colonoscopic biopsy (n=3) at Asan Medical Center from 2010 to 2016 were used. All samples had MSI results that were confirmed by standard PCR analysis, NGS test, or both. We used five markers (BAT25, BAT26, D2S123, D5S346, and D17S250) recommended by the National Cancer Institute (NCI) for the standard PCR MSI analysis. Diagnosis of the MSI status through NGS was carried out as described previously [
7]. Hematoxylin and eosin–stained slides from FFPE were reviewed by two pathologists (M.L and J.K.) for tumor distribution and cellularity. The areas of the slide presenting the highest tumor cellularity were marked, and tumor cellularity was evaluated by rough estimation of the proportion of tumor cell nuclei relative to non-neoplastic cell nuclei. In order to determine the minimum required tumor purity, the tumor area was marked in various ways so that various proportions of tumor nuclei could be included in the analytes in two cases.
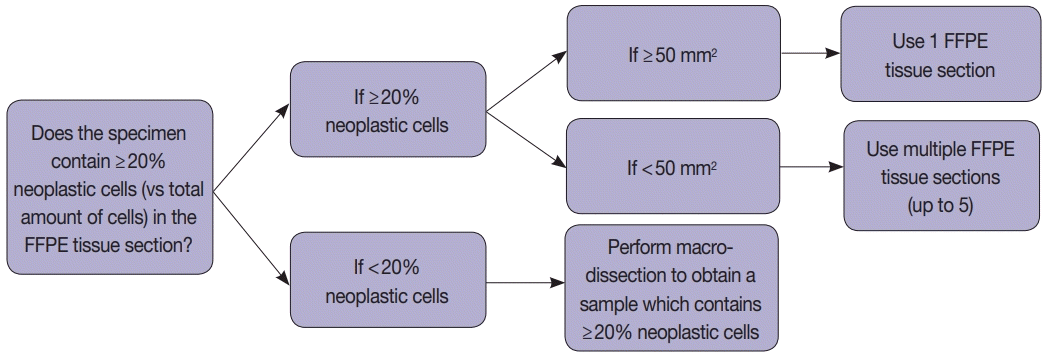
Tissue handling was done according to the instructions provided by the manufacturers (
Fig. 1). Briefly, macro-dissection was done if the tumor cellularity was less than 20%, and whole cut sections were used if it was more than 20% (
Fig. 1). We obtained 5-µm FFPE tissue sections: 1 section when the tumor area is equal to or greater than 50 mm
2 and 2–5 sections when the tumor area is less than 50 mm
2 depending on the tumor surface area (
Fig. 1). In most cases, macro-dissection was not required. The scraped FFPE tissue fragments were directly put into the cartridge.
Fig. 1.
Sample processing protocol depending on the tumor purity and tissue surface area. FFPE, formalin-fixed paraffin-embedded.

The Idylla MSI assay
The Idylla MSI assay (Biocartis NV, Mechelen, Belgium) was performed as described previously [
9]. Briefly, the scraped FFPE tissue fragments were loaded directly onto a designated cartridge and the device was run for approximately 150 minutes. Detection of these specific targets was performed using fluorescently labeled molecular beacons after PCR amplification. These beacons melt differentially from the wild type or mutated amplicons with increasing temperature. The MSI test–specific software, namely, Test Type Package, automatically checked the validity of the measured fluorescence profiles: the presence of specific PCR amplicons resulted in biomarker-specific fluorescence profiles. Next, a powerful pattern-recognition algorithm trained on several thousands of profiles calculated a probability score (MSI score) for any given valid biomarker-specific profile. This MSI score per biomarker expressed the probability of a pattern being wild type or mutant. As such, MSI biomarkers were scored individually and reported as “No mutation detected,” “Mutation detected” or “Invalid.” The Idylla MSI assay uses seven biomarkers, namely
ACVR2A,
BTBD7,
DIDO1,
MRE11,
RYR3,
SEC31A, and
SULF2. Those biomarkers were chosen based on their short length that is advantageous for probe-based PCR, stability over different cancer types and ethnicities, minimized false positives and negatives, and diagnostic performance optimized for the test platform. A test result was considered valid if ≥5 out of 7 MSI biomarkers show valid amplified signals. The tumors were classified into two categories. High frequency of MSI (MSI-H) was defined if two or more of the seven markers were positive, and microsatellite stable (MSS) if less than two of seven markers were positive.
Determination of limit of detection
We randomly selected four cases that were confirmed as MSI-H by standard PCR analysis and determined the limit of detection through stepwise tumor marking that results in varying degrees of tumor cellularity. As shown in
Supplementary Fig. S1, we designated the tumor areas with varying degrees of nonneoplastic cells so that the approximate tumor cellularity is 80%, 60%, 40%, 20%, or 10%, respectively. In three cases, samples with approximately 5% and/or 2% tumor purity were also analyzed (
Supplementary Fig. S1).
Statistical analysis
We compared the results of Idylla MSI assay with the combined gold standard MSI results for the 105 cases. The sensitivity, specificity, positive predictive value, negative predictive value, and accuracy of Idylla MSI assay were calculated for each test in comparison with the combined gold standard.
Ethics statement
Written informed consent was obtained from all patients and this study was approved by the Institutional Review Board (protocol number: 2018-1518).
RESULTS
Idylla MSI assay
All 115 cases showed “valid” results. Thirteen out of 115 cases (11.3%) were classified as MSI-H (
Table 1). Mutations were detected in at least four of seven marker genes in these 13 MSI-H cases (
Table 1). Among the MSS samples, most cases did not reveal any mutation in the seven markers; however, a particular case (case No. 71) showed
MRE11 mutation on Idylla MSI assay but the assay categorized it as MSS because the probability score was below the threshold.
Table 1.
A detailed list of the mutated marker genes in the MSI-H cases that were determined by the Idylla MSI assay
|
Case No. |
Biomarker
|
|
ACVR2A
|
BTBD7
|
DIDO1
|
MRE11
|
RYR3
|
SEC31A
|
SULF2
|
Mutant marker/Total makers |
|
4 |
Mut |
Mut |
Mut |
Mut |
Mut |
No |
Mut |
6/7 |
|
28 |
Mut |
Mut |
No |
Mut |
No |
No |
Mut |
4/7 |
|
52 |
Mut |
Mut |
Mut |
No |
Mut |
Mut |
Mut |
6/7 |
|
54 |
Mut |
Mut |
Mut |
No |
Mut |
Mut |
Mut |
6/7 |
|
57 |
Mut |
Mut |
Mut |
Mut |
No |
Mut |
Mut |
6/7 |
|
65 |
Mut |
Mut |
Mut |
Mut |
Mut |
Mut |
No |
6/7 |
|
104 |
No |
No |
Mut |
Mut |
Mut |
No |
Mut |
4/7 |
|
113 |
Mut |
Mut |
Mut |
Mut |
Mut |
Mut |
Mut |
7/7 |
|
117 |
Mut |
Mut |
Mut |
Mut |
Mut |
Mut |
Mut |
7/7 |
|
118 |
Mut |
Mut |
Mut |
Mut |
Mut |
Mut |
Mut |
7/7 |
|
120 |
Mut |
Mut |
Mut |
Mut |
No |
Mut |
No |
5/7 |
|
124 |
Mut |
Mut |
Mut |
No |
No |
Mut |
Mut |
5/7 |
|
132 |
Mut |
No |
Mut |
Mut |
Mut |
Mut |
Mut |
6/7 |

Validation of the MSI status through standard PCR analysis, NGS, or both
Standard PCR analysis for MSI using the five markers recommended by NCI were carried out in the 108 cases except for the seven cases that were not available for tissue analysis. Of the 108 cases, 89 cases were MSS, five cases were low frequency of MSI (MSI-L), and 14 cases were MSI-H. The NGS test for MSI was performed on all 115 cases, and three of them failed during the library preparation step of NGS analysis. Of the 112 cases with valid NGS results, 101 cases were MSS while 11 cases were MSI-H. The results of standard PCR analysis and the results of NGS test results for MSI were completely concordant in all 105 cases with both test results, except for the cases without available tissue material for standard PCR analysis and those that failed the NGS test (
Table 2). We combined the results of standard PCR analysis and NGS test and thereafter used the combined results as a gold standard. For standard PCR results, MSI-L results were considered MSS.
Table 2.
Correlation between standard PCR analysis and NGS for MSI
|
Colorectal carcinoma (n = 105) |
Standard PCR analysis
|
Total |
|
MSS |
MSI-L |
MSI-H |
|
NGS |
|
|
|
|
|
Stable |
89 |
5 |
0 |
94 |
|
Unstable |
0 |
0 |
11 |
11 |
|
Total |
89 |
5 |
11 |
105 |

Comparison between Idylla MSI assay and the gold standard MSI results
We compared the results of Idylla MSI assay with the combined gold standard MSI results for the 105 cases with both results of standard PCR analysis and NGS test. Idylla MSI assay achieved 99.05% accuracy (104 of 105), 100% sensitivity (11 of 11), 98.94% specificity (93 of 94), 91.67% positive predictive value (11 of 12), and 100% negative predictive value (93/93) for the detection for MSI (
Table 3). Only one case (case No. 28) showed discordant result between the Idylla MSI assay and the gold standard result. MSS result was confirmed by both standard PCR fragment analysis (
Fig. 2A) and NGS (
Fig. 2C), but the Idylla MSI assay called it MSI-H.
Fig. 2.
Details of three cases with discrepant results between the Idylla microsatellite instability (MSI) assay and the standard polymerase chain reaction (PCR) test. (A–C) Case No. 28, MSI-high (MSI-H) according to the Idylla MSI assay. Standard PCR MSI test is consistent with microsatellite stable (MSS) (A). MSH2 and MLH1 expression is shown in immunochemical staining (B), and next-generation sequencing (NGS) results show MSS pattern (C). (D–F) Case No. 131, MSS (0/7) according to the Idylla MSI assay. Two markers show instability in standard PCR MSI test (arrows, D), and MSH6 and PMS2 protein expressions are shown (E). Repeated NGS shows MSS pattern (F). (G, H) Case No. 133, MSS (0/7) according to the Idylla MSI assay. Two markers show instability in standard PCR MSI test (arrows, G), and MSH6 and PMS2 expressions are shown (H). Repeated NGS analysis was failed. Mutation load, the number of total somatic mutations detected by our NGS panel; TMB, tumor mutation burden, the inferred number of somatic mutations per megabase; I index, the number of insertion or deletion mutations divided by the number of all mutations; IHC, immunohistochemistry
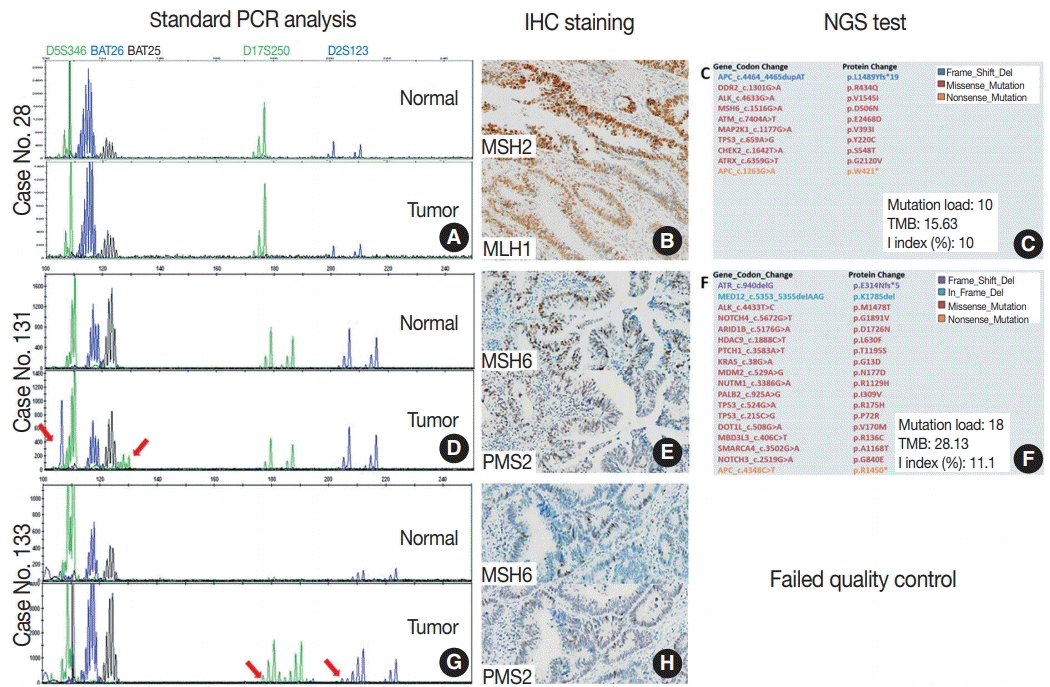

Table 3.
Comparison between the Idylla MSI assay results and the combined gold standard MSI results
|
Colorectal carcinoma (n = 105) |
Idylla MSI assay
|
Total |
|
MSS |
MSI-H |
|
Combined gold standard MSI |
|
|
|
|
MSS |
93 (98.9) |
1 (1.1) |
94 |
|
MSI-H |
0 |
11 (100) |
11 |
|
Total |
93 |
12 |
105 |

Further evaluation of the cases that failed the NGS test
To investigate whether the Idylla MSI assay can accept a wider range of samples than NGS, we selected three cases that were confirmed as MSI-H in standard PCR test but failed in our NGS test. Among them, two cases showed instability on only two microsatellite markers (case Nos. 131 and 133) (
Fig. 2D–
H), while the other case (case No. 132) showed instability in all five markers (
Table 4). Among them, only one case (case No. 132) showed concordant results and the other two showed discordant results. Interestingly, the two cases with discordant results were the cases where instability was observed in two markers in the standard PCR analysis. To resolve these discrepancies, additional immunohistochemical staining for MSH2, MSH6,
MLH1, and PMS2 were performed and standard PCR analysis and NGS was repeated. Although repeated PCR MSI tests were not helpful due to the high background noise band (data not shown), protein expression of all the MMR proteins was preserved in the two cases (
Fig. 2B,
E), suggesting that those two cases were likely MSS. Furthermore, the second attempt of NGS was successful in case No. 131 and it showed MSS pattern although the tumor mutation burden was relatively high (28.13/Mb) (
Fig. 2F). The second attempt of NGS was not successful in case No. 133. As expected, the MSI-H phenotype of the case with concordant standard PCR and Idylla MSI results was confirmed through the loss of MSH2 and MSH6 expression (
Table 4).
Table 4.
Analysis of the cases that failed in the NGS test
|
Case No. |
Standard PCR analysis No. 1 |
Repeated NGS analysis |
Immunohistochemical staining
|
Idylla MSI assay |
|
MSH2 |
MSH6 |
MLH1 |
PMS2 |
|
131 |
MSI-H, 2/5 |
MSS |
Positive |
Positive |
Positive |
Positive |
MSS, 0/7 |
|
132 |
MSI-H, 5/5 |
Not done |
Negative |
Negative |
Positive |
Positive |
MSI-H, 6/7 |
|
133 |
MSI-H, 2/5 |
Failed |
Positive |
Positive |
Positive |
Positive |
MSS, 0/7 |

Limit of detection analysis of the Idylla MSI assay
To determine the minimum tumor purity that does not affect the assay results, we selected a few MSI-H samples and macrodissected each sample in such a way that various levels of tumor purity could be achieved. Case No. 118 was successfully categorized as MSI-H when the tumor purity was lowered down to 10% (
Table 5). Likewise, case Nos. 104 and 113 were successfully categorized as MSI-H even in samples with lower tumor purity of 2%. However, in case No. 52, which was diagnosed as MSI-H based on the presence of mutations in six out of seven markers, no single mutation was detected in all seven biomarkers when the tumor cellularity was 2%. In this particular case, there were several foci of lymphoid aggregates which may further dilute the tumor cells (
Supplementary Fig. S1E).
Table 5.
MSI detection ability of the Idylla MSI assay according to variable tumor cellularity
|
Case No. |
Tumor cellularity (%) |
Marker gene
|
Mutant marker/Total makers |
|
ACVR2A
|
BTBD7
|
DIDO1
|
MRE11
|
RYR3
|
SEC31A
|
SULF2
|
|
118 |
80 |
Mut |
Mut |
Mut |
Mut |
Mut |
Mut |
Mut |
7/7 |
|
60 |
Mut |
Mut |
Mut |
Mut |
No |
Mut |
Mut |
6/7 |
|
40 |
Mut |
Mut |
Mut |
Mut |
No |
Mut |
Mut |
6/7 |
|
20 |
Mut |
Mut |
Mut |
Mut |
No |
Mut |
Mut |
6/7 |
|
10 |
Mut |
Mut |
Mut |
Mut |
No |
Mut |
Mut |
6/7 |
|
104 |
80 |
No |
No |
Mut |
Mut |
No |
No |
Mut |
3/7 |
|
60 |
No |
No |
Mut |
Mut |
Mut |
No |
Mut |
4/7 |
|
40 |
No |
No |
Mut |
Mut |
No |
No |
Mut |
3/7 |
|
20 |
No |
No |
Mut |
Mut |
No |
No |
Mut |
3/7 |
|
10 |
No |
No |
Mut |
Mut |
No |
No |
No |
2/7 |
|
5 |
No |
No |
Mut |
Mut |
No |
No |
Mut |
3/7 |
|
2 |
No |
No |
Mut |
Mut |
No |
No |
No |
2/7 |
|
113 |
80 |
Mut |
Mut |
Mut |
Mut |
Mut |
Mut |
Mut |
7/7 |
|
2 |
Mut |
No |
Mut |
No |
Mut |
No |
Mut |
4/7 |
|
52 |
80 |
Mut |
Mut |
Mut |
No |
Mut |
Mut |
Mut |
6/7 |
|
2 |
No |
No |
No |
No |
No |
No |
No |
0/7 |

DISCUSSION
In this study, the Idylla MSI assay demonstrated excellent diagnostic performance when we tested the fully automated kit with 115 CRC samples with rigorously confirmed MSI results. In addition, the test successfully detected MSI-H status in samples with very low tumor purity (down to 10%), although the tumor purity estimates were inevitably crude. Our findings suggest that the Idylla MSI assay could be used in the clinic to determine MSI status, at least for CRC samples, and that macro-dissection may not be required in most CRC cases. Since the Idylla MSI assay does not require DNA extraction and reagent loading steps, the hands-on time could be greatly reduced.
The identification of MMR deficiency is important not only for the identification of the risk for Lynch syndrome but also for the appropriate treatment approach and prognosis for sporadic tumors [
10]. MMR deficiency can be diagnosed by PCR fragment analysis, NGS, and MMR protein IHC test [
8]. MMR IHC staining is used as a first-line screening method and is used as a good surrogate marker for MSI, but heterogeneity has previously been well described [
10,
11]. PCR fragment analysis has been widely used as a standard method and NGS-based MSI detection is now increasing, but both methods are time-consuming and labor-intensive. The Idylla MSI assay offers a fully automated workflow from direct FFPE tissue input to a simple final report (
Supplementary Fig. S2) and achieved a short turnaround time (about 150 minutes) and minimal hands-on time (less than 2 minutes). When we simulated actual hands-on time in maximum throughput environment (8 samples/run for the Idylla MSI assay, 32 samples/run for the standard MSI PCR assay, and 24 samples/run for the NGS in Illumina NextSeq platform), the hands-on time per sample was 30 seconds for the Idylla MSI assay, 5 minutes for the standard MSI PCR test, and 120 minutes for the NGS. Thus, the Idylla MSI assay may significantly save time and manpower. The downside of the Idylla MSI assay, however, is that only one sample can be processed in each instrument unit at a time. Thus, to increase sample throughput, multiple instruments are required (up to 8 instruments per one console, according to the manufacturer). Requirements for multiple instruments and usage of one sophisticated cartridge per one sample may make this test to be more expensive than the standard PCR assay.
Although the Idylla MSI assay results were concordant with gold standard results in most cases, there was one false-positive case (case No. 28). The gold standard result was MSS but the Idylla MSI assay categorized it as MSI-H based on the presence of mutations in four genes such as ACVR2A, BTBD7, MRE11, and SULF2. Since the MSS result was confirmed by both standard PCR fragment analysis and NGS, it is unlikely that the gold standard result was incorrect. Furthermore, cross-contamination or sample swapping was rigorously excluded through repeated analysis and DNA fingerprinting that is included in our NGS assay. It could be that the particular tumor harbored somatic mutations in the four genes, irrespective of MMR deficiency. Indeed, one MSS tumor (case No. 71) showed MRE11 mutation on the Idylla MSI assay but the assay categorized it as MSS because the total probability score was below the threshold. Thus, rare false MSI-H results may be possible in a particular MSS tumor harboring mutations in the genes covered by the Idylla MSI assay.
Most molecular techniques involving homogenized samples, in which the tumor cells and non-neoplastic cells are mixed, require tumor marking with subsequent macro-dissection to ensure sufficient tumor purity. Regarding this, the direct use of unstained FFPE tissues cut without macro-dissection could maximize the benefits of the fully automated Idylla MSI assay. In this study, we found that most FFPE samples may not require macro-dissection because the test reliably detected MSI-H in samples with low tumor purity (approximately 10%). However, since the ability to detect MSI-H was not perfectly consistent with the very low tumor purity (approximately 2%), we propose that the minimum required tumor purity would be approximately 10%. In addition, the age of the FFPE tissue material may not cause any significant problem in most cases because the test was successful in FFPE samples aged up to 9 years.
We also tested whether the Idylla MSI assay might accept poor quality samples better than the NGS assay by testing samples that were diagnosed as MSI-H with the standard PCR test but failed in our NGS analysis. All the Idylla MSI assay results were “valid” which means successful amplification of the adequate number of marker genes, suggesting that the Idylla MSI assay may accept some “difficult” samples better than NGS. Furthermore, MMR protein IHC study and repeated NGS analysis suggested that the Idylla MSI assay may sometimes be more accurate than the standard PCR MSI test because the two samples, which were MSS on the Idylla MSI assay but were MSI-H on the standard PCR test, showed unequivocal expression of four MMR proteins and a relatively low number of unstable markers (only two unstable markers out of five markers) in the standard PCR MSI test. In addition, in one of the two samples, MSS could be confirmed through repeated NGS analysis. However, the full validation of the performances of the Idylla MSI assay might be limited because of a relatively small size of the MSI-H sample group in our study.
In summary, the Idylla MSI assay is fast, accurate, and reliable, and thus might be clinically applicable. The fully automated workflow may offer a significant reduction in time and labor although the cost for consumables or cartridge is higher than that of the standard MSI PCR test. In addition, colonoscopic biopsy specimens in the case of initially metastatic disease may be easily processed because this test does not require a matched normal tissue sample.





 PDF
PDF Citation
Citation Print
Print




 XML Download
XML Download