This article has been
cited by other articles in ScienceCentral.
Abstract
Background
Single staining is commonly performed for practical pathologic diagnoses. However, this method is limited in its ability to specify cellular morphology and immunophenotype and often requires consumption of limited tissue. This study aimed to describe an optimized protocol for multiple in situ hybridization (ISH) and immunohistochemistry (IHC).
Methods
The quality of multistaining was evaluated by carefully changing each step of ISH and IHC in an angioimmunoblastic T-cell lymphoma (AITL) case on a Ventana BenchMark XT automated immunostainer. The optimized protocols were also performed using another immunostainer and in 15 cases of five Epstein-Barr virus (EBV)–associated malignancies using formalin-fixed paraffin-embedded tissue.
Results
The quality of various ISH-IHC staining protocols was semi-quantitatively evaluated. The best EBV-encoded RNA (EBER)-ISH/double IHC staining quality, equivalent to single staining, was obtained using the following considerations: initial EBER-ISH application, use of protease and antigen retrieval reagent (cell conditioning 1 [CC1] treatment time was minimized due to impact on tissue quality), additional baking/ deparaffinization not needed, and reduced dilution ratio and increased reaction time for primary antibody compared with single immunostaining. Furthermore, shorter second CC1 treatment time yielded better results. Multiple staining was the best quality in another immunostainer and for different types of EBV-associated malignancies when it was performed in the same manner as for the Ventana BenchMark XT as determined for AITL.
Conclusions
EBER-ISH and double IHC could be easily used in clinical practice with currently available automated immunostainers and adjustment of reagent treatment time, dilution ratio, and antibody reaction time.
Keywords: Protocol optimization of multistaining, In situ hybridization, Immunohistochemistry, Epstein-Barr virus-associated malignancy, Formalin-fixed paraffin-embedded tissue
Pathologic diagnosis is usually based on a comprehensive analysis of biomarker expression along with morphological features of lesions. An integrated analysis of the expression pattern of various biomarkers may be a decisive factor in pathologic diagnosis. For example, in lymphoproliferative lesions of childhood, whether atypical cells express Epstein-Barr virus (EBV)-encoded RNA (EBER), and whether they express CD4, CD8, CD56, and/or CD30 may play a key role in diagnosis [
1]. Treatment plans for patients may be determined by the expression of specific biomarkers, such as with CD20 expression for rituximab therapy in diffuse large B-cell lymphoma (DLBCL) [
2]. In pathologic practices,
in situ hybridization (ISH) or immunohistochemistry (IHC) is usually performed to detect genetic material or the expression of protein antigens because of their effectiveness and efficiency. The number of biomarkers evaluated for diagnosis has increased, increasing the need for minimal tissue consumption for the stateof-the-art ancillary tests, especially in small biopsies [
3]. However, until recently, ISH and IHC have usually been stained with a single marker per slide.
Multistaining can be presented as a way to both reduce tissue consumption and easily identify the cell type expressing biomarkers and patterns of expression. The benefits of multiple IHC or immunofluorescence staining have already been demonstrated in several cancer diagnostic strategies [
4-
6]. In cases where multiple ISH and IHC procedures are performed, the possibility of tissue damage and antigen loss becomes high, and it becomes difficult to precisely evaluate biomarker expression and morphology of the tissue at a practical level [
7]. Most automated immunostainers automate all staining steps from deparaffinization to visualization, thereby minimizing errors that can occur during manual work and greatly improving accuracy [
8]. However, there are few studies on well-established staining methods for multistaining with ISH and IHC using automated immunostainers.
This study aimed to devise an optimized protocol for multiple ISH and IHC staining on automated immunostainers. The quality of multistaining was evaluated by carefully changing each step of ISH and IHC with formalin-fixed paraffin-embedded (FFPE) tissues of EBV-associated malignancies.
MATERIALS AND METHODS
Specimens
A case of angioimmunoblastic T-cell lymphoma (AITL) with sufficient resected lymph node tissue and confirmed EBV infection at primary diagnosis was selected as the representative specimen for testing ISH and IHC staining conditions. A total of 15 EBV-associated malignancies were further used to validate the optimal multistaining protocol. These 15 samples included three EBV-positive DLBCL, three extranodal natural killer/T-cell lymphomas, three classical Hodgkin lymphomas (cHL) of mixed cellularity type, three AITLs, and three EBV-positive gastric carcinomas with lymphoid stroma (EBV-GCLS). Four EBV-negative malignancies including peripheral T-cell lymphoma and gastric adenocarcinoma were stained with the optimized protocol as controls. All specimens were FFPE tissues and each was cut to a thickness of 4 μm.
EBER-ISH and IHC
EBER-ISH and IHC for protein antigens were performed using the Ventana BenchMark XT automatic immunostainer following the manufacturer’s recommendations (Ventana Medical Systems, Tucson, AZ, USA). EBER-ISH was done with an ISH iView Blue Detection Kit. Protein removal and nucleic acid exposures used ISH protease 2 (#780-4148) or ISH protease 3 (#780-4149), respectively, and the reaction times were varied. Counterstaining was not performed.
Details of the primary antibodies used in IHC and probes used in ISH are summarized in
Supplementary Table S1 . Two different coloring agents, 3,3'-diaminobenzidine (DAB) and new fuchsin, were used for double ISH-IHC or triple ISH-IHC staining either alone or in combination. For color development with DAB, the OptiView DAB Detection Kit (hereafter, OptiView kit) was used. Baking and deparaffinization steps were either performed or omitted. When indicated, baking was carried out at 65°C for 10 minutes. For heat-induced epitope retrieval (HIER), treatment time of cell conditioning 1 (CC1) was varied. The dilution ratio of primary antibody was varied taking into consideration the dilution ratio used in single IHC, and the reaction time was varied at 37°C. Counterstaining was performed or omitted as indicated. For color development with new fuchsin, the UltraView Universal Alkaline Phosphatase Red Detection Kit (hereafter, UltraView kit) was used. CC1 treatment time was varied. Dilution ratio and reaction times of primary antibodies were also varied. Counterstaining was performed or omitted as indicated.
EBER-ISH and IHC were also performed using another immunostainer, the Leica Bond III automated immunostainer (Leica Biosystems, Melbourne, Australia). EBER-ISH was performed with the Bond polymer refine detection kit, fluorescein-conjugated EBER oligonucleotide probe (#PB0589) and a mouse monoclonal anti-fluorescein antibody (#AR0833). Procedures were carried out in the same manner as on the Ventana equipment.
Evaluation of the quality of multistaining protocols
The quality of ISH-IHC staining protocols was semi-quantitatively scored by a pathologist (H.G.) and a medical laboratory scientist (J.K.J.) according to the following criteria relative to the quality of a single stain of EBER-ISH or counterstain (
Supplementary Fig. S1A–
S1C): score 1, no expression (<1% of cells for which target antigen expression is expected) and scarce preservation of cell or nucleus shape; score 2, rare expression (1% to <10%) and poor preservation; score 3, minimal expression (10% to <50%) and bare preservation; score 4, moderate expression (50% to <75%) and fair preservation; Score 5, abundant expression (≥75%) and good preservation. In the case of IHC, intensity of the staining and suitability of the color development position, i.e., location of expression, were evaluated from 1 to 5 points.
The quality of ISH, IHC, and hematoxylin counterstain was separately evaluated and scored. The scores were weighted based on the comparison between the results of single staining and staining according to the nominal protocol. The greater the change in the state of staining from the previous step to the later step, the greater the weight was assigned to the later step, except for the counterstain. More specifically, ISH, the first procedure of multistaining, was given a weight of 0.15 because it was minimally affected by various factors that reduce the quality of the staining. The intensity and location of IHC were given a weight of 0.30 and 0.35, respectively, because those parameters were more useful in achieving the purpose of immunostaining to evaluate the expression of a targeted antigen, while counterstaining with hematoxylin was given a weight of 0.20. The total score was added to the score of each item to which weights were applied and the maximum was 5.
Ethics statement
Exemption from informed consent after de-identification of samples was approved by the Institutional Review Board of Asan Medical Center (2017-0522).
RESULTS
Multistaining according to nominal protocol
A simple repeat protocol according to the single staining protocols of ISH and IHC was termed the nominal protocol, P1. When EBER-ISH and CD20- or CD3-IHC staining was sequentially performed on the same slide tissue section of AITL, EBER expression was maintained at the same level as single staining, but CD20 and CD3 were poorly expressed, denaturation of the tissue was so severe that it was difficult to identify the shape of the cells and the quality of the counterstain was also poor (
Supplementary Fig. S1D,
S1E). As expected, EBER-ISH was not expressed at all using the reverse P1 protocol, i.e., performing IHC first and then ISH, with the same AITL case (
Supplementary Fig. S2A,
S2B).
Optimization of double ISH-IHC staining protocol
EBER-ISH and CD20-IHC double staining was performed under various conditions with the same tissue used for P1. Double ISH-IHC staining was conducted to tune optimal conditions for five factors, i.e., ISH-protease, CC1, additional baking/deparaffinization, primary antibody dilution and primary antibody reaction time. These factors were considered to affect the quality of multistaining and each protocol was designated from P2-1 to DS-4 as summarized in
Tables 1 and
2. The reaction time of ISH-protease 2 was varied to minimize the deterioration of CD20-IHC and counterstain and to maintain the quality of EBER-ISH. ISH-protease 3 was set to 12 minutes and ISH-protease 2 was varied to 4, 8, or 12 minutes under CC1 treatment fixed at 32 minutes. Compared with those of the single stain, quality of CD20-IHC was approximately 60% (score 0.9) and 47% (0.7) improved, respectively, and counterstain quality was the same, but the EBER-ISH quality was deteriorated (0.45) with ISH-Protease 3. Thus, ISH-Protease 3 was discarded. In protocols using ISH-protease 2, protease reaction time did not show any deterioration or enhancement in the quality of EBER-ISH, but longer reaction times of ISH-protease worsened CD20-IHC and the counterstain. In P2-2 with the shortest ISH-protease treatment, EBER-ISH quality was not deteriorated and counterstaining showed good results. However, the score for CD20 IHC was only about 42.7% (1.6/3.25) of the single stain. The total score was 3.35, the highest among P2 protocols (
Fig. 1A).
Fig. 1.
Multistaining using each protocol for optimization with angioimmunoblastic T-cell lymphoma tissue. (A) P2-2 (Epstein-Barr virus–encoded RNA [EBER], purple; CD20, brown). (B) P3-2 (EBER, purple; CD20, brown). (C) P4-2 (EBER, purple; CD20, brown). (D, E) DS-2 (EBER, purple; CD20, brown). (F, G) TS-4 (EBER, purple; CD20, red; CD3, brown). (H, I) double EBER in situ hybridization and CD20 immunohistochemistry staining on a Leica Bond III immunostainer (EBER, brown; CD20, red).
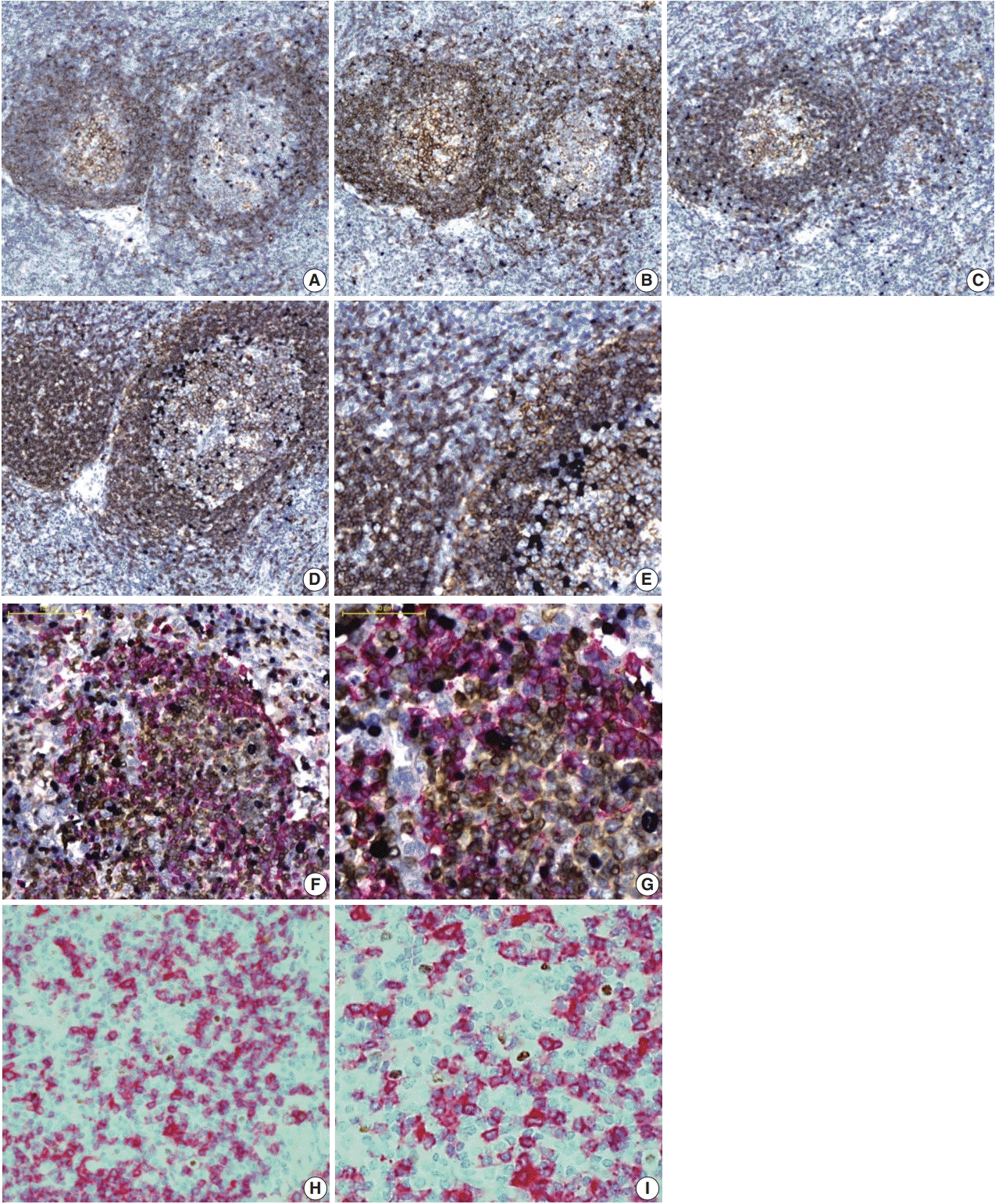

Table 1.
Nominal and optimization protocols of double staining for EBER-ISH and CD20-IHC on a Ventana BenchMark XT immunostainer
|
Protocol |
First procedure (EBER)
|
Second procedure (CD20)
|
|
Protease (min) |
Baking (min) |
Deparaffinization |
CC1 (min) |
Ab dilution |
Ab incubation (min) |
|
P1 |
Protease 2: 8 |
10 |
Selected |
32 |
1:200 |
32 |
|
Optimization protocol for ISH protease |
|
P2-1 |
Protease 3: 12a
|
ND |
ND |
32 |
1:200 |
32 |
|
P2-2 |
Protease 2: 4a
|
ND |
ND |
32 |
1:200 |
32 |
|
P2-3 |
Protease 2: 8a
|
ND |
ND |
32 |
1:200 |
32 |
|
P2-4 |
Protease 2: 12a
|
ND |
ND |
32 |
1:200 |
32 |
|
Optimization protocol for CC1 |
|
P3-1 |
Protease 2: 4 |
ND |
ND |
8a
|
1:200 |
32 |
|
P3-2 |
Protease 2: 4 |
ND |
ND |
16a
|
1:200 |
32 |
|
P3-3 |
Protease 2: 4 |
ND |
ND |
32a
|
1:200 |
32 |
|
P3-4 |
Protease 2: 4 |
ND |
ND |
40a
|
1:200 |
32 |
|
Optimization protocol for baking and deparaffinization |
|
P4-1 |
Protease 2: 4a
|
10a
|
Selecteda
|
8a
|
1:200 |
32 |
|
P4-2 |
Protease 2: 4a
|
10a
|
Selecteda
|
16a
|
1:200 |
32 |
|
P4-3 |
Protease 2: 4a
|
10a
|
Selecteda
|
32a
|
1:200 |
32 |
|
P4-4 |
Protease 2: 8a
|
10a
|
Selecteda
|
8a
|
1:200 |
32 |
|
P4-5 |
Protease 2: 8a
|
10a
|
Selecteda
|
16a
|
1:200 |
32 |
|
P4-6 |
Protease 2: 8a
|
10a
|
Selecteda
|
32a
|
1:200 |
32 |
|
P4-7 |
Protease 2: 12a
|
10a
|
Selecteda
|
8a
|
1:200 |
32 |
|
P4-8 |
Protease 2: 12a
|
10a
|
Selecteda
|
16a
|
1:200 |
32 |
|
P4-9 |
Protease 2: 12a
|
10a
|
Selecteda
|
32a
|
1:200 |
32 |
|
Optimization protocol for antibody dilution and incubation time |
|
DS-1 |
Protease 2: 4 |
ND |
ND |
16a
|
1:200a
|
60a
|
|
DS-2 |
Protease 2: 4 |
ND |
ND |
16a
|
1:200a
|
120a
|
|
DS-3 |
Protease 2: 4 |
ND |
ND |
16a
|
1:100a
|
60a
|
|
DS-4 |
Protease 2: 4 |
ND |
ND |
16a
|
1:100a
|
120a
|

Table 2.
Scores of the optimization protocols for multistaining on a Ventana BenchMark XT immunostainer
|
Protocol |
EBER |
Protein expression
|
Hematoxylin |
Total score |
CD20
|
CD3
|
|
Intensity |
Location |
Intensity |
Location |
|
P1 |
0.75 |
0.3 |
0.35 |
ND |
ND |
0.2 |
1.6 |
|
Optimization protocol of double staining for ISH protease |
|
P2-1 |
0.45 |
0.9 |
0.7 |
ND |
ND |
1 |
3.05 |
|
P2-2 |
0.75a
|
0.9a
|
0.7a
|
ND |
ND |
1a
|
3.35a
|
|
P2-3 |
0.75 |
0.9 |
0.7 |
ND |
ND |
0.4 |
2.75 |
|
P2-4 |
0.75 |
0.6 |
0.35 |
ND |
ND |
0.2 |
1.9 |
|
Optimization protocol of double staining for CC1 |
|
P3-1 |
0.75 |
0.6 |
0.7 |
ND |
ND |
1 |
3.05 |
|
P3-2 |
0.75a
|
0.9a
|
0.7a
|
ND |
ND |
1a
|
3.35a
|
|
P3-3 |
0.75 |
0.9 |
0.7 |
ND |
ND |
0.8 |
3.15 |
|
P3-4 |
0.75 |
0.9 |
1.05 |
ND |
ND |
0.6 |
3.3 |
|
Optimization protocol of double staining for baking/deparaffinization |
|
P4-1 |
0.75 |
0.6 |
0.7 |
ND |
ND |
1 |
3.05 |
|
P4-2 |
0.75a
|
0.9a
|
0.7a
|
ND |
ND |
1a
|
3.35a
|
|
P4-3 |
0.75 |
0.9 |
0.7 |
ND |
ND |
0.8 |
3.15 |
|
P4-4 |
0.75 |
0.3 |
0.35 |
ND |
ND |
0.6 |
2 |
|
P4-5 |
0.75 |
0.6 |
0.7 |
ND |
ND |
0.6 |
2.65 |
|
P4-6 |
0.75 |
0.9 |
0.7 |
ND |
ND |
0.4 |
2.75 |
|
P4-7 |
0.75 |
0.3 |
0.35 |
ND |
ND |
0.4 |
1.8 |
|
P4-8 |
0.75 |
0.3 |
0.35 |
ND |
ND |
0.4 |
1.8 |
|
P4-9 |
0.75 |
0.3 |
0.35 |
ND |
ND |
0.2 |
1.6 |
|
Optimization protocol of double staining for antibody dilution and incubation time |
|
DS-1 |
0.75 |
0.9 |
1.4 |
ND |
ND |
1 |
4.05 |
|
DS-2 |
0.75 |
1.2 |
1.4 |
ND |
ND |
1 |
4.35 |
|
DS-3 |
0.75 |
0.9 |
1.4 |
ND |
ND |
1 |
4.05 |
|
DS-4 |
0.75a
|
1.5a
|
1.75a
|
ND |
ND |
1a
|
5a
|
|
Optimization protocol of triple staining with CD3-IHCb
|
|
TS-1 |
0.75 |
1.5 |
1.75 |
0.9 |
1.05 |
1 |
3.7 |
|
TS-2 |
0.75a
|
1.5a
|
1.75a
|
1.5a
|
1.75a
|
1a
|
5a
|
|
TS-3 |
0.75 |
1.5 |
1.75 |
1.5 |
1.4 |
0.8 |
4.45 |
|
TS-4 |
0.75 |
1.5 |
1.75 |
1.5 |
1.05 |
0.6 |
3.9 |

CC1 reaction time (P3 protocols) was varied to 8, 16, 32, and 40 minutes. The quality of CD20-IHC staining increased with longer CC1 reaction time, but the counterstain quality gradually decreased. Among P3 protocols, the quality of staining was best for P3-2 with a total score of 3.35 (
Fig. 1B). Next, the effects of baking and additional deparaffinization on the quality of staining were evaluated. Nine P4 protocols were designed by setting the baking time to 10 minutes and varying ISH-protease 2 and CC1 reaction times. In all cases, baking and deparaffinization did not deteriorate or improve the quality of staining (
Fig. 1C).
Finally, the effects of dilution ratio and timing of primary antibody on the quality of multistaining were evaluated (DS protocols). Based on the optimal dilution ratio and time for CD20 single staining, the dilution ratio of CD20 antibody was halved from 1:200 to 1:100 and/or the reaction time was doubled from 60 to 120 minutes. As a result, the DS-4 protocol with a dilution factor of 1:100 of CD20 primary antibody and a reaction time of 120 minutes resulted in a total score of 5 by summing up the scores of EBER-ISH (0.75), CD20-IHC (3.25), hematoxylin (1) while maintaining a level of quality equivalent to the result of a single stain for each procedure in all evaluation factors (
Fig. 1D,
E).
Optimization of triple stain of EBER-ISH and double IHC
Based on optimized conditions with EBER and CD20 staining, CD3-IHC triplicate staining was performed under various conditions with the same AITL tissue. The quality of ISH and double IHC staining was evaluated by changing the same five factors as well as the ISH-IHC staining. Among them, CC1 reaction time of CD3-IHC was varied in the same manner as CD20-IHC (
Table 3). Consequentially, even when CD3 antibody was added, the best results were obtained when staining was performed in the same manner as ISH-IHC staining, which maintained the same quality of staining as single stains with each probe or primary antibody (
Fig. 1F,
G).
Table 3.
Optimization of triple staining for EBER-ISH and CD20- and CD3-IHC staining on a Ventana BenchMark XT immunostainer
|
Protocol |
First procedure (EBER-ISH)
|
Second procedure (CD20-IHC)
|
Third procedure (CD3-IHC)
|
|
Protease (min) |
CC1 (min) |
Ab incubation (min) |
Dilution |
CC1 (min) |
Ab incubation (min) |
Dilution |
|
TS-1 |
4 |
16 |
120 |
1:100 |
8a
|
120 |
1:100 |
|
TS-2 |
4 |
16 |
120 |
1:100 |
16a
|
120 |
1:100 |
|
TS-3 |
4 |
16 |
120 |
1:100 |
32a
|
120 |
1:100 |
|
TS-4 |
4 |
16 |
120 |
1:100 |
46a
|
120 |
1:100 |

To further evaluate the applicability of the optimized protocol, ISH-IHC staining was performed on a Leica Bond III automated immunostainer with each case of AITL, cHL, and EBV-GCLS. As with a Ventana immunostainer in all three cases, the best staining quality was obtained when EBER-ISH was first applied and Enzyme I protease treatment time was minimized. In more detail, EBER was fully expressed following treatment with protease K for 10 minutes in both single EBER-ISH and ISH-IHC, and background staining occurred with protease K treatment for 20 minutes or more and when the incubation time of the EBER probe was increased from 45 minutes in single EBER-ISH to 3 hours in ISH-IHC. Antibody dilution ratio was halved and antibody reaction time was doubled compared with single immunostaining conditions (
Fig. 1H,
I).
Validation of the optimized protocols in EBV-associated and non-associated malignancies
The optimized multi-EBER-ISH and double IHC staining protocols were validated in 15 EBV-associated (3 cases per cancer type) and four EBV-non-associated (2 cases per cancer type) malignancies. Staining conditions were the same as in the TS-4 protocol, and the dilution ratio of each antibody was concentrated to half the dilution of the single stain while reaction time was doubled (
Table 4).
Table 4.
Multi-EBER-ISH and IHC staining using optimized protocols in EBV-associated malignancies using a Ventana BenchMark XT immunostainer
|
EBER CD20 CD3 |
EBER CD20 CD56 |
EBER CD3 CD15 |
EBER CD30 CD3 |
EBER CK CD3 |
|
First procedure, XT INFORM Probe iVIEW Blue v3 |
|
|
|
|
|
|
1. Deparaffinization |
S |
S |
S |
S |
S |
|
2. ISH Protease 2 (min) |
4 |
4 |
4 |
4 |
4 |
|
3. INFORM EBER |
S |
S |
S |
S |
S |
|
Second procedure, XT OptiView DAB IHC v4 |
|
|
|
|
|
|
4. CC1 (min) |
16 |
16 |
16 |
16 |
16 |
|
5. Pre-primary peroxidase inhibit |
S |
S |
S |
S |
S |
|
6. Ab incubation time (min) |
120 |
120 |
120 |
120 |
120 |
|
Ab dilution |
1:100 |
1:100 |
1:100 |
1:50 |
1:100 |
|
Third procedure, XT UltraView RED v3 |
|
|
|
|
|
|
7. CC1 (min) |
20 |
20 |
20 |
20 |
20 |
|
8. Ab incubation time (min) |
120 |
120 |
120 |
120 |
120 |
|
Ab dilution |
1:100 |
1:25 |
1:100 |
1:100 |
1:100 |
|
9. Hematoxylin 2 (min) |
16 |
16 |
16 |
16 |
16 |
|
10. Bluing (min) |
4 |
4 |
4 |
4 |
4 |

As showed in
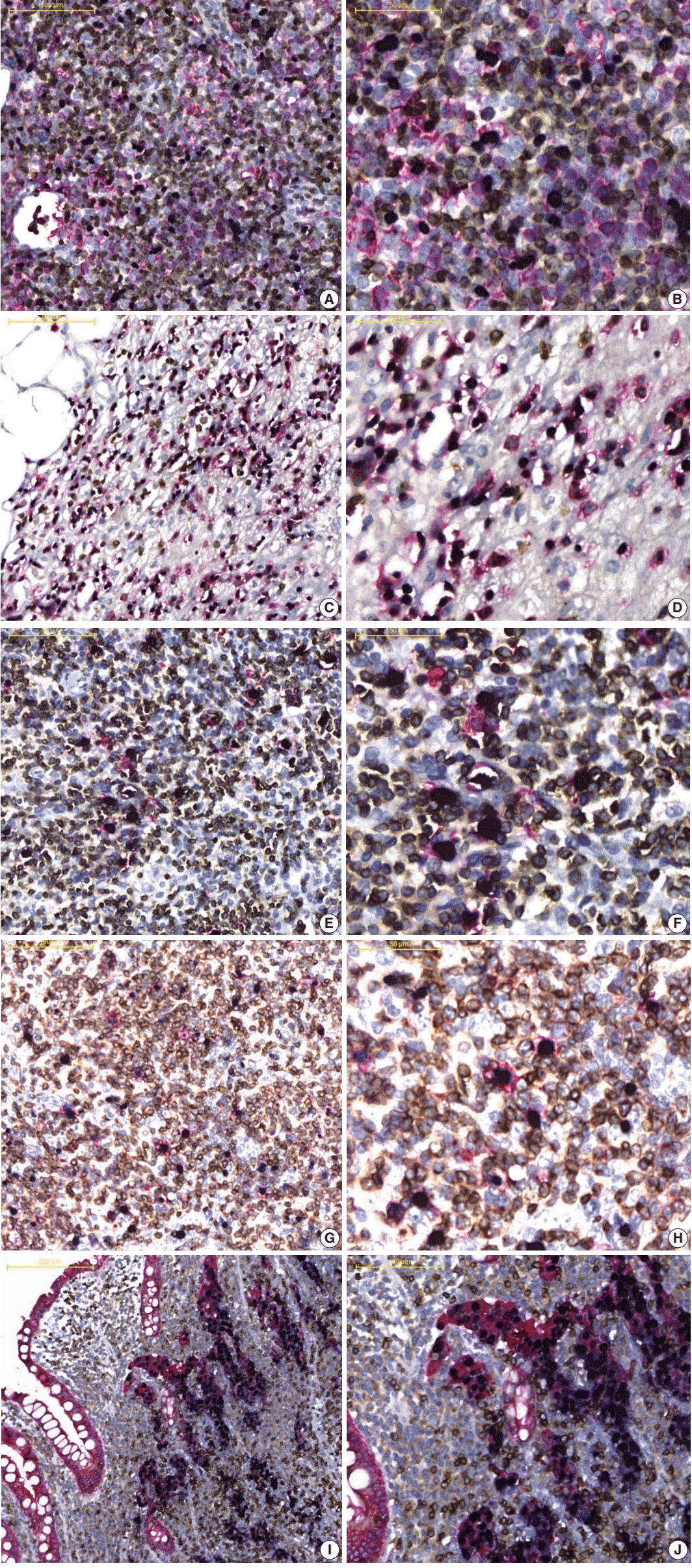
Fig. 2A and
B, in the EBV-positive DLBCL, (1) EBER-ISH and CD20 (red)-/CD3 (DAB)-IHC and (2) EBER-ISH and CD20 (red)-/CD30 (DAB)-IHC staining was performed to evaluate whether EBV-infected cell types were identifiable. CD20-positive cells expressing a red membrane were large with atypical nuclei and diffusely proliferated. These cells were frequently observed with dark blue color nuclei, and the surrounding small lymphocytes were admixed with red- or brown-expressing cells. These colors indicated that EBV-infected cells were not small T or B cells, but rather large atypical B cells. In addition, in EBER-ISH and CD20-/CD30-IHC staining, some EBER- and CD20-positive cells were observed to be reddish brown. When staining results were taken together, EBV-positive DLBCL were found to be neoplastic B cells infected with EBV, and some of these cells expressed CD30. Similarly, it was easy to determine which biomarker was expressed on EBV-infected cells compared to single staining in other types of EBV-associated malignancies (
Fig. 2C–
J,
Supplementary Fig. S3). Furthermore, for all tested EBV-associated malignancies, the staining quality of each biomarker and counterstain was nearly identical to that of a single stain. In contrast, EBER was never expressed but the staining quality was well preserved in all EBV-non-associated malignancies (
Supplementary Fig. S4).
Fig. 2.
Application of the optimized multistaining protocol to various Epstein-Barr virus (EBV)–associated malignancies. (A, B) EBV-positive diffuse large B-cell lymphoma (Epstein-Barr virus–encoded RNA [EBER], purple; CD20, red; CD3, brown). (C, D) Extranodal natural killer/T-cell lymphoma, nasal type (EBER, purple; CD56, red; CD3, brown). (E, F) Classic Hodgkin lymphoma stroma (EBER, purple; CD15, red; CD3, brown). (G, H) Angioimmunoblastic T-cell lymphoma stroma (EBER, purple; CD30, red; CD3, brown). (I, J) EBV-positive gastric carcinoma with lymphoid stroma (EBER, purple; cytokeratin, red; CD3, brown).

DISCUSSION
This study aimed to identify an optimized multistaining protocol using automated immunostainers and commercially available kits for ISH and IHC to make multiple staining easy to use in pathologic practice. Recently, a proper assessment of the expression of many biomarkers in diagnoses has been become increasingly important. In particular, it is essential to accurately determine the immunophenotype of various biomarkers and morphology of cells in diagnosing many benign or malignant diseases associated with EBV infection [
9]. However, the immunophenotyping of atypical cells as well as the EBER-expressing cells is challenging and the various molecular tests frequently needed consume a lot of tissue. Thus, we selected EBV-associated malignancies to be representative examples for establishing optimized multistaining protocols.
We determined that ISH should be performed before IHC during multiple staining. When IHC was administered first, EBER-ISH was not stained, which could be attributed to the complete degradation of mRNA during IHC. Additional baking/deparaffinization was not needed for antigen retrieval during IHC, but all of the primary antibodies used in this study required HIER for antigen retrieval. If proteolytic-induced epitope retrieval (PIER) is required for restoring antigenicity, such as with the primary antibody for epidermal growth factor receptor [
10], it may be necessary to determine PIER reaction time during IHC with multistaining because this protease would have already been applied in ISH. It was expected that staining quality might be worse due to tissue damage when PIER reaction time was the same in multi-ISH/IHC stain as in single IHC. Although a definite mechanism for HIER has yet to be established, it is presumed that the calcium ion of the HIER solution plays a role of opening or closing the cross linkage between proteins through a chelation reaction with a covalent ring-forming metal substance [
11]. In double IHC, temporarily opened protein cross-linking after HIER is restored to the state before HIER after a certain period of time, and only one HIER is required when the two primary antibodies simultaneously react. However, we needed additional HIER because the primary antibodies were applied in sequence in double IHC.
The OptiView kit and the UltraView kit for IHC are 3-step and 2-step multimer methods, respectively. Because the sensitivity of antigen detection is higher with the OptiView kit than the UltraView kit, the dilution ratio of some antibodies needed to be reduced when used with the UltraView kit. Thus, we obtained better staining with primary antibodies of higher sensitivity using the UltraView kit, while those with lower sensitivity were stained using the OptiView kit when double IHC was performed. In addition, the 3-step multimer method, which maintained good staining quality even when CC1 treatment time was short, was first applied, and then the 2-step multimer method was applied while adjusting appropriate CC1 treatment time. The development of various coloring agents for 3-step multimers would provide more flexibility in protocol design and result in better quality multi-immunostaining [
12].
ISH-protease and HIER time were found to be the most important parameters for successful multiple staining. The longer the treatment time of ISH-protease, the more severe the loss of antigen. Longer HIER led to worse tissue condition and poor quality hematoxylin counterstaining, making it difficult to identify tissue architecture and cell morphology. In the case of multiple ISH/IHC staining, the protease and HIER time proposed in this study could be used to achieve the same quality of staining as single ISH and/or IHC.
Because three staining procedures were sequentially performed for EBER-ISH and double IHC, the kits needed to be replaced at the end of each procedure, and the turnaround time increased. Nevertheless, double EBER-ISH/IHC and triple EBER-ISH/ IHCs could be completed within the same day or take up to half a day. Because FFPE tissues and three independent kits were used for triple staining in each procedure, the staining procedures for each marker could be easily added. For example, if double or triple staining was required while reading an EBER-ISH stained slide, the coverslip of the slide could be removed and stained with other biomarkers such as CD20, CD30, and CD15.
In conclusion, EBER-ISH and double IHC could be easily used in pathologic practice by adjusting the treatment time of the reagents as well as the dilution ratio and reaction time of the primary antibodies, as suggested by the described optimized protocols for currently available automated immunostainers. If more coloring agents are commercialized in the future, more biomarker expression could be evaluated on one slide. In addition, evaluation of the type of cells expressing a particular biomarker and the relevance of multiple biomarkers would be more accurate, while minimizing tissue loss for further ancillary studies.






 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download