Abstract
Liquid biopsy for detection of mutation from circulating tumor DNA is a new technology which is attractive in that it is non-invasive. Epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKI) is an effective first line drug for advanced non-small cell lung cancer patients who harbor activating EGFR mutation. During the course of treatment, resistance against TKI arises which can be contributed to EGFR T790M mutation in about 50–60% of patients. Third generation TKI may overcome the resistance. In patients who cannot undergo tissue biopsy due to variable reasons, liquid biopsy is an excellent alternative for the detection of EGFR T790M mutation. However, this relatively novel method requires standardization and vigorous quality insurance. Thus, a standard set of guideline recommendations for liquid biopsy for EGFR mutation testing suitable for the Korean medical community is necessary. In this article, we propose a set of provisional guideline recommendations that was discussed and approved by the Cardiopulmonary Pathology Study Group of the Korean Society of Pathologists.
After the discovery of activating epidermal growth factor receptor (EGFR) gene mutation, EGFR tyrosine kinase inhibitors (TKI) became the first line of treatment in advanced non-small cell lung cancer (NSCLC) with mutated EGFR [1-3]. These EGFR TKIs such as gefitinib, erlotinib, and afatinib show consistently better response rate and prolonged progression-free survival in EGFR mutant NSCLC patients [1-3]. However, most patients receiving EGFR TKI treatment may develop acquired resistance [4-6]. Although various mechanisms are involved in this resistance, secondary T790M mutation of EGFR gene illustrates 50%–60% of the resistance [7,8]. A recently developed third generation TKIs can effectively target T790M, and so it is very critical to detect this mutation in patients who has developed acquired resistance against first- or second-line EGFR TKIs [9-11].
Liquid biopsy is an emerging tool that detects genetic changes in circulating tumor DNA (ctDNA) shed from the tumor cells [12-14]. Recently, Cobas EGFR mutation test V2 (Roche, Indianapolis, IN, USA) has been approved by Food and Drug Administration (FDA) for the detection of EGFR mutations from the blood of NSCLC patients [15]. Although this non-invasive technique is fascinating and promising, it is still a developing method which needs further improvements. Hence, it is necessary to have guidelines for its usage. Korean cardiopulmonary study group has prepared the first guideline of EGFR mutation detection in blood for clinicians and pathologists who actively take part in the diagnosis and treatment of lung cancer.
Liquid biopsy for the detection of EGFR mutation can play many roles in cancer diagnostics [12-14,16,17]. Patients diagnosed with lung adenocarcinoma harboring EGFR mutation will be the first candidates when they develop resistance against first-line TKIs. Especially, when the tumor is too small or located in a challenging region to be sampled, liquid biopsy can be a good alternative [14-18]. Patients with poor performance status can also benefit from this technique.
Sample collection and processing is a critical step in liquid biopsy. Since ctDNA is rapidly degraded by the nuclease in blood and contaminated by genomic DNA from blood cells, it is essential to separate plasma from the sample [13,14]. The routine venipuncture technique will be sufficient to collect blood from the patients. The sample collection tube should be chosen considering each institution’s setting. Conventional ethyldiaminetetraaceticacid (EDTA) tube can be used if the samples are processed without delay [19,20]. Recently, specialized tubes for delaying degradation of ctDNA are commercially available [19,20]. The tube from Streck (Omaha, NE, USA) has been the most widely used collection tube. Roche diagnostics and Qiagen have also marketed specialized tubes. According to a study [19], conventional EDTA tube and Streck tube do not show much difference in their performance when samples are processed within 6 hours. When incubated longer in EDTA tube, cell-free DNA may be released from the blood cells, and EDTA will hinder the polymerase chain reaction (PCR) [20]. Tubes from Roche and Qiagen showed similar performance, and they are slightly better than Streck tube [20]. Specialized tubes can sustain sample quality for several days at room temperature before processing further (Table 1).
Before ctDNA extraction, blood should be processed into plasma through double centrifugation. Plasma samples are better than serum samples, which can be contaminated by DNA released from immune cells [13]. Since a small amount of ctDNA is present in plasma, isolation is a critical step in the process for saving tumor DNA. Several commercial kits for isolation are available in the market (Table 2) [21,22]. These are manual, semiautomatic, and fully automatic. Manual protocol uses column-based method while semi-automatic instrument works with magnetic beads. Previous studies showed variable results depending on the extraction kits, though they all had similar performances [21,22]. The technician’s skill and protocol optimization may be one of the critical factors for yielding better ctDNA. Table 1 summarizes commercial ctDNA extraction kits.
High sensitivity detection methods are required to detect EGFR mutations from liquid samples. Kits for detecting mutations have been developed and are commercially available [23-25]. Each kit requires different quality and amount of DNA (Table 3). They depend on real time PCR technology with their own variations. Roche Cobas uses real time PCR with Taqman like probe and Qiagen has released ARMS based kits, Therascreen EGFR RGQ. Another PCR based technique uses peptide nucleic acid clamping and Panamutyper (Panagene, Daejeon, Korea). The Roche and Qiagen systems use their own PCR machine from Roche and Qiagen while Panamutyper can run on any qualified PCR machines. The number of mutations these kits can detect are different; however, together they include exon 19 deletion, T790M and L858R. Currently, only Roche kit has acquired FDA approval. The most important element of these kits is how sensitively and specifically they can detect mutations in liquid samples. There are certain studies to evaluate their performance and report sensitivities ranging from 62% to 67.5% and specificities ranging from 88% to 97% [26-29]. In the ASSESS study, these three kits showed high specificity, however, sensitivity was equal to or less than 75% [25]. For T790M, sensitivity was 41% and 29% for Cobas and Therascreen, respectively, and specificity was 100% for both kits from the patients enrolled in AURA trial [10]. Therefore, deciding the best kit will depend on the laboratory’s choice with consideration of their requirements. Features of these products are summarized in Table 3. Other platforms using digital PCR and next generation sequencing are still far from widespread use in clinical setting [24].
Once liquid biopsy for detecting T790M mutation is done, the reports should contain the following information: pathologic number, age, sex, hospital unit number, sample source, requesting physician, requesting department, adequacy for testing (amount of DNA extracted), receipt day, report day, storage tube, methodology used, exons tested and associated range of detectable mutations, mutation status, comments, testing technician, and corresponding pathologist. Since the patients already have sensitizing EGFR mutation, it is recommended to include the type of original EGFR alteration and previous histologic diagnosis.
Since liquid biopsy technique has not been validated yet, vigorous quality assurance is necessary. Although there is no recommended program for external quality assessment (EQA), one pilot trial for EGFR testing in blood is ongoing in Germany [30]. Another program for BRAF and KRAS is also being conducted [31]. Since patient derived standard sample is difficult to store and distribute, artificial sample mimicking the real one can be used instead [30,31]. We are in the process of developing Korean EQA program.
Performance and interpretation of liquid biopsy require broad knowledge in lung cancer pathology. Pathologists have an important role in the diagnosis and management of cancer and thus can interpret liquid biopsy results in conjunction with the histologic diagnosis, previous status of EGFR-activating mutation, and clinical situation. The liquid biopsy in lung cancer is usually performed in patients whose previous EGFR mutation status has been known. The sole purpose of this technique is to detect a T790M mutation responsible for TKI resistance. Unlike tissue specimens, in which the pathologists can determine the percentage of tumor cells, it is extremely difficult to estimate whether the blood sample contains a sufficient amount of tumor DNA. If the sample is adequate, the test generally finds the original EGFR-activating mutation, which may act as an internal control for the presence of ctDNA [13]. When it has not detected any EGFR-activating mutation including previously existing one or reported mutations other than the preexisting ones or T790M, pathologists should be able to interpret the result. In the former, test should be repeated because the samples might have been degraded and contain insufficient ctDNA. In the latter, the newly emerged mutation, in the presence of newly developed lesion, may indicate a metachronous primary tumor. The communication between pathologists, clinicians, and radiologists is important for further diagnosis and management of cancer. Moreover, lung adenocarcinoma undergoes frequent transformation into small cell carcinoma when it is treated with TKI, while maintaining the original EGFR mutation [7,8,32]. Recommended interpretation is suggested in Table 4.
EGFR mutation testing performed with blood or other liquid sample is a non-invasive method, which can be more widely adopted. Laboratories must get familiar with liquid samples and develop their own protocols to handle these specimens. They can choose appropriate sample tubes, extraction kits, detection methods, and other instruments. They should select the most suitable combination in accordance with their requirements, unless the detection kits indicate specific methods and instruments [31]. Although sensitivity of tissue biopsy is higher than liquid biopsy, both are far from perfection and T790M mutation can be detected only in one of the two methods. Reportedly, allele fraction of T790M mutation tends to correlate with treatment efficacy of osimertinib [33]. Therefore, absence of T790M in tumor tissue while it is detected in plasma might reflect low allele frequency and lead to poor response. Therefore, the two methods are complementary to each other and should be selected according to each patient’s condition (Fig. 1).
Liquid biopsy is a promising method, which is safe and convenient. Before more experiences and data are accumulated, liquid biopsy should be performed with great caution. There are a few steps in liquid biopsy, which can produce false negative or false positive results. Interpretation requires profound knowledge of lung cancer including diagnosis, treatment, and prognosis. However, in debatable cases, discussion between pathologists, physicians, and radiologists is critical. This method will soon play a major role in early diagnosis, monitoring of treatment, and detection of minimal residual disease. Currently, it cannot replace the conventional pretreatment tissue diagnosis [14]. It is important to validate and improve the performance of this technique before it is widely used in clinical practice. Liquid biopsy performed in EGFR has provided a platform for determining gene mutations in KRAS, ALK, PI3CA, and BRAF as well.
Notes
Author contributions
Conceptualization: DHS, HSS, TJK, HSP, YLC, WSK, LK, SHC, JSS, HJK, JHH, CHL, GKL, SJJ.
Data curation: DHS.
Formal analysis: DHS, HSS, TJK.
Investigation: DHS, HSP, YLC, WSK, LK.
Methodology: DHS, SHC, JSS, HJK, JHH.
Project administration: JHH, CHL, GKL, SJJ.
Writing—original draft: DHS.
Writing—review & editing: DHS, HSS, TJK, HSP, YLC, WSK, LK, SHC, JSS, HJK, JHH, CHL, GKL, SJJ.
REFERENCES
1. Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004; 350:2129–39.

2. Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004; 304:1497–500.
3. Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009; 361:947–57.

4. Balak MN, Gong Y, Riely GJ, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006; 12:6494–501.

5. Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015; 21:560–2.

6. Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011; 3:75ra26.

7. Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011; 3:75ra26.

8. Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013; 19:2240–7.
9. Goss G, Tsai CM, Shepherd FA, et al. Osimertinib for pretreated EGFR Thr790Met-positive advanced non-small-cell lung cancer (AURA2): a multicentre, open-label, single-arm, phase 2 study. Lancet Oncol. 2016; 17:1643–52.
10. Yang JC, Ahn MJ, Kim DW, et al. Osimertinib in pretreated T790Mpositive advanced non-small-cell lung cancer: AURA study phase II extension component. J Clin Oncol. 2017; 35:1288–96.

11. Ballard P, Yates JW, Yang Z, et al. Preclinical comparison of osimertinib with other EGFR-TKIs in EGFR-mutant NSCLC brain metastases models, and early evidence of clinical brain metastases activity. Clin Cancer Res. 2016; 22:5130–40.
12. Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008; 14:985–90.

13. Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017; 17:223–38.

14. Sholl LM, Aisner DL, Allen TC, et al. Liquid biopsy in lung cancer: a perspective from members of the Pulmonary Pathology Society. Arch Pathol Lab Med. 2016; 140:825–9.

15. Cobas EGFR Mutation Test v2, PMA 150047. FDA summary of safety and effectiveness data [Internet]. Silverspring: US Food and Drug Administration;2016. [cited 2018 Jan 2]. Available from: https://www.accessdata.fda.gov/cdrh_docs/pdf15/P150047B.pdf.
16. Mino-Kenudson M. Cons: Can liquid biopsy replace tissue biopsy?: the US experience. Transl Lung Cancer Res. 2016; 5:424–7.
17. Ilie M, Hofman P. Pros: can tissue biopsy be replaced by liquid biopsy? Transl Lung Cancer Res. 2016; 5:420–3.
18. Mock TS, Carbone DP, Hirsch FR. IASLC atlas of EGFR testing in lung cancer. Aurora: International Association for the Study of Lung Cancer;2017.
19. Alidousty C, Brandes D, Heydt C, et al. Comparison of blood collection tubes from three different manufacturers for the collection of cell-free DNA for liquid biopsy mutation testing. J Mol Diagn. 2017; 19:801–4.

20. Kang Q, Henry NL, Paoletti C, et al. Comparative analysis of circulating tumor DNA stability In K3EDTA, Streck, and CellSave blood collection tubes. Clin Biochem. 2016; 49:1354–60.

21. Fleischhacker M, Schmidt B, Weickmann S, et al. Methods for isolation of cell-free plasma DNA strongly affect DNA yield. Clin Chim Acta. 2011; 412:2085–8.

22. Sorber L, Zwaenepoel K, Deschoolmeester V, et al. A comparison of cell-free DNA isolation kits: isolation and quantification of cellfree DNA in plasma. J Mol Diagn. 2017; 19:162–8.
23. Vendrell JA, Mau-Them FT, Beganton B, Godreuil S, Coopman P, Solassol J. Circulating cell free tumor DNA detection as a routine tool for lung cancer patient management. Int J Mol Sci. 2017; 18:E264.
24. Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: a cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer. 2015; 90:509–15.
25. Reck M, Hagiwara K, Han B, et al. ctDNA determination of EGFR mutation status in European and Japanese patients with advanced NSCLC: the ASSESS study. J Thorac Oncol. 2016; 11:1682–9.
26. Bernabe R, Hickson N, Wallace A, Blackhall FH. What do we need to make circulating tumour DNA (ctDNA) a routine diagnostic test in lung cancer? Eur J Cancer. 2017; 81:66–73.
27. Qiu M, Wang J, Xu Y, et al. Circulating tumor DNA is effective for the detection of EGFR mutation in non-small cell lung cancer: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2015; 24:206–12.
28. Luo J, Shen L, Zheng D. Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: a systematic review and meta-analysis. Sci Rep. 2014; 4:6269.

29. Wu Y, Liu H, Shi X, Song Y. Can EGFR mutations in plasma or serum be predictive markers of non-small-cell lung cancer? A meta-analysis. Lung Cancer. 2015; 88:246–53.
30. Fassunke J, Ihle MA, Lenze D, et al. EGFR T790M mutation testing of non-small cell lung cancer tissue and blood samples artificially spiked with circulating cell-free tumor DNA: results of a round robin trial. Virchows Arch. 2017; 471:509–20.
31. Haselmann V, Ahmad-Nejad P, Geilenkeuser WJ, et al. Results of the first external quality assessment scheme (EQA) for isolation and analysis of circulating tumour DNA (ctDNA). Clin Chem Lab Med. 2018; 56:220–8.

Fig. 1.
Proposed diagnostic algorithm for the detection of epidermal growth factor receptor (EGFR) T790M mutation. NSCLC, nonsmall cell lung cancer; TKI, tyrosine kinase inhibitor; CTx, chemotherapy.
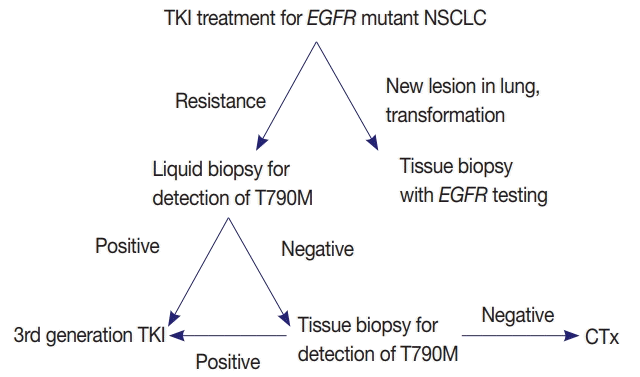
Table 1.
Comparison of specialized tubes for collection of ctDNA
| Company | Trade name | Volume (mL) | Temperature (°C) | Storage duration (day) |
|---|---|---|---|---|
| Streck | cfDNA BCT | 10 | 6–37 | 14 |
| Roche | Cell-Free DNA Collection Tube | 8.5 | 18–25 | 7 |
| Qiagen | PAXgene Blood ccfDNA Tube | 10 | 18–25 | 7 |
Table 2.
Commercially available ctDNA extraction kits
| Company | Trade name | Method | Automation |
|---|---|---|---|
| ThermoFisher | MagMAX | Magnetic beads | Semiauto |
| Promega | Maxwell RSC | Magnetic beads | Semiauto |
| Roche | Cobas | Silica membrane | Manual |
| Qiagen | QIAamp | Silica membrane | Semiauto |
Table 3.
EGFR mutation detection kits in plasma
Table 4.
Recommended interpretation of EGFR mutation test from blood




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download