Abstract
Background
Protein phosphatase magnesium-dependent 1δ (PPM1D) is a p53-induced serine/threonine phosphatase, which is overexpressed in various human cancers. A recent study reported that a mutation in the PPM1D gene is associated with poor prognosis in brainstem gliomas. In this study, we evaluated the utility of PPM1D as a prognostic biomarker of adult supratentorial diffuse astrocytic and oligodendroglial tumors.
Methods
To investigate PPM1D protein expression, mRNA expression, and copy number changes, immunohistochemistry, RNAscope in situ hybridization, and fluorescence in situ hybridization were performed in 84 adult supratentorial diffuse gliomas. We further analyzed clinical characteristics and overall survival (OS) according to PPM1D protein expression, and examined its correlation with other glioma biomarkers such as isocitrate dehydrogenase (IDH) mutation, and p53 expression.
Results
Forty-six cases (54.8%) were PPM1D-positive. PPM1D expression levels were significantly correlated with PPM1D transcript levels (p= .035), but marginally with PPM1D gene amplification (p=.079). Patients with high-grade gliomas showed a higher frequency of PPM1D expression than those with low-grade gliomas (p <.001). Multivariate analysis demonstrated that PPM1D expression (hazard ratio [HR], 2.58; p=.032), age over 60 years (HR, 2.55; p=.018), and IDH1 mutation (HR, 0.18; p=.002) were significantly independent prognostic factors; p53 expression had no prognostic significance (p=.986). The patients with tumor expressing PPM1D showed a shorter OS (p=.003). Moreover, patients with tumor harboring wild-type IDH1 and PPM1D expression had the worst OS (p<.001).
Gliomas are the most common tumors of the brain and spinal cord, and account for the majority of brain cancer-related deaths [1,2]. Molecular-profiling studies have reported characteristic genetic alterations related with different gliomas. These biomarkers were subsequently incorporated in the 2016 World Health Classification (WHO) Classification of Tumours of the Central Nervous System (revised 4th edition) [3-9]. Mutations in the isocitrate dehydrogenase 1 (IDH1) and IDH2 genes occur in 70%–80% of grade II/III gliomas and most secondary glioblastomas [10]. Co-deletion of 1p/19q is typically related with tumors of the oligodendroglial lineage and is closely associated with IDH mutations [11]. Alterations in the p53 and its pathway genes occur in 78% of glioblastomas, and are thought to promote progression to high-grade malignancy [12,13].
Protein phosphatase magnesium-dependent 1δ (PPM1D), also known as wild-type p53-induced phosphatase 1, is a member of the protein phosphatase type 2C and p53 target gene family and occurs in response to various stresses [14-18]. Activated PPM1D directly dephosphorylates Chk1, Chk2, p38 mitogen-activated protein kinase, uracil DNA glycosylase, MDM2, H2AX, and p53, suggesting that it acts as a homeostatic regulator [19-22]. The PPM1D gene is frequently amplified/overexpressed in various human cancers, all of which rarely carry a p53 mutation [23-29]. Recent studies have reported C-terminal truncating alterations in the PPM1D gene, which enhance the functional ability of PPM1D. PPM1D mutation was identified in 18.2% of pediatric brainstem gliomas, which were mutually exclusive with p53 mutations detected in 57.6% of the same tumors. Moreover, PPM1D mutation was known to be associated with poor prognosis in pediatric brainstem gliomas [30-32].
In this study, we intended to evaluate the utility of PPM1D expression as a prognostic biomarker of adult supratentorial diffuse astrocytic and oligodendroglial tumors. Other clinical characteristics were also analyzed to explore the relationship between the tumors and PPM1D expression. Moreover, we investigated brain lower grade gliomas (WHO grade II and III) from The Cancer Genome Atlas (TCGA) data to clarify the clinical effects according to the genetic alterations of PPM1D.
We retrospectively reviewed data for 109 patients diagnosed with diffuse astrocytic and oligodendroglial tumors according to the 2016 WHO Classification of Tumours of the Central Nervous System (revised 4th edition) from August 2013 to July 2015 from the archives of the Department of Pathology at Asan Medical Center. Eighty-four patients were enrolled after excluding four cases of adult or pediatric brainstem glioma, one case of adult cerebellar glioma, and 20 cases without sufficient tissue. The medical records of 84 patients were reviewed, including sex, age, tumor locations, diagnoses, molecular parameters, treatments, and survival outcomes. This study adhered to the guidelines established by the Declaration of Helsinki and was approved by the Institutional Review Board of Asan Medical Center (2015-0151). Informed consents were obtained from all individual participants included in the study.
Tissue microarrays (TMAs) consisting of two cylindrical cores (3-mm) from formalin-fixed, paraffin-embedded tissues obtained from surgically resected or stereotactic biopsy specimens were constructed using a Quick-Ray Manual Tissue Microarrayer (UT06, Unitma, Belrose, NSW, Australia). Immunohistochemistry (IHC) of 4-μm paraffin section of TMA blocks was performed using a Benchmark automatic immunostaining device (Ventana Medical Systems, Tucson, AZ, USA). The slides were incubated with primary antibodies against PPM1D (1:100, 2804D1a, Abcam, Cambridge, UK) and p53 (1:1,500, DO.7, DAKO, Glostrup, Denmark). IHC for PPM1D was scored as follows: 0, no reactivity or nuclear and cytoplasmic reactivity in less than 5% of the tumor cells; 1, reactivity in 5% to less than one-third of the tumor cells; 2, reactivity in one-third to two-thirds of the tumor cells; and 3, reactivity in more than two-thirds of the tumor cells [33]. Immunopositivity was determined to optimize the cutoff point for patient survival to dichotomize the PPM1D expression data using the X-Tile software [34]. Scores of 0 and 1 indicated negative results, and scores of 2 and 3 indicated positive results. p53 immunoreactivity was also scored using this method.
The genomic region spanning codon 132 of IDH1 was amplified using the polymerase chain reaction with the following primer set: 5'-TGAGAAGAGGGTTGAGGAGTTC-3' (forward) and 5'-CACATACAAGTTGGAAATTTCTGG-3' (reverse). The genomic region was then sequenced with the forward primer using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster, CA, USA).
PPM1D transcript was measured with the RNAscope assay using the Hs-PPM1D probe (602231; Advanced Cell Diagnostics, Hayward, CA, USA). Hybridization signals were amplified and visualized with the RNAscope 2.5 HD reagent kit-RED (322350, Advanced Cell Diagnostics). PPM1D mRNA molecules, shown as red spots, were quantified via light microscopy by manual counting. The signals per cell were divided into four groups according to the manufacturer’s semi-quantitative scoring guideline: score 0, no staining or less than 1 dot in 10 cells; score 1, 1–3 dots in a cell; score 2, 4–9 dots in a cell; and score 3, ≥10 dots in a cell or the presence of gene clusters in ≥ 10% of the tumor cells. Based on a cutoff value of 1.5 determined by using the X-Tile software, scores of 2 and 3 were considered to indicate high PPM1D mRNA expression, and scores of 0 and 1 indicated no and low PPM1D mRNA expression, respectively.
Copy number status of the PPM1D gene was evaluated via fluorescence in situ hybridization (FISH) using the HYBrite denaturation/hybridization system (Abbott Molecular, Chicago, IL, USA). The PPM1D FISH probes in red and chromosome 17 control probes (CEP17) in green were purchased from Empire Genomics (Buffalo, NY, USA). Specimens that showed a PPM1D/CEP17 ratio ≥ 3 were considered positive for PPM1D amplification, and those with a PPM1D/CEP17 ratio > 1 but < 3 in ≥ 10% of tumor cells were defined as relative copy gain.
The independent t test, chi-square test, and Fisher exact test were carried out to assess the association between PPM1D protein expression and clinicopathological characteristics. To determine the relationships among PPM1D alterations, we conducted a chi-square test and Fisher exact test. The Cox proportional hazards regression model was used to assess the dependency of survival duration on predictor variables. To estimate survival rates and compare survival distribution, we used the Kaplan-Meier method and log-rank test, respectively. All statistical analyses were performed using the R software (ver. 3.3.2, the R Foundation for Statistical Computing, Vienna, Austria). Any p-value < .05 was assumed to indicate a statistically significant difference.
A total of 84 patients with adult supratentorial diffuse astrocytic and oligodendroglial tumors were analyzed. The clinicopathological findings are summarized in Table 1. The 84 cases consisted of 26 glioblastomas, IDH-wildtype (31.0%), 16 anaplastic oligodendrogliomas, IDH-mutant and 1p/19q-codeleted (19.0%), 12 oligodendrogliomas, IDH-mutant and 1p/19q-codeleted (14.3%), and 10 diffuse astrocytomas, IDH-mutant (11.9%). The majority (89.7%, 61/68) of the patients with high-grade gliomas or diffuse astrocytomas (IDH-mutant and wildtype) underwent surgery with adjuvant chemotherapy or chemoradiation therapy. Those with oligodendrogliomas, IDH-mutant and 1p/19q-codeleted were treated with surgery without any additional therapy (100%, 12/12). Over half of the enrolled patients showed PPM1D expression (54.8%, 46/84) and p53 expression (54.8%, 46/84) in IHC. Among the 36 patients (42.9%) with IDH1 mutations, 35 had R132H mutation and 1 had R132S mutation.
IHC for PPM1D was performed in 84 cases of adult diffuse astrocytic and oligodendroglial tumors (Fig. 1A). The relationships between PPM1D expression and clinicopathological characteristics are summarized in Table 1. The patients’ characteristics were not significantly different according to PPM1D expression except for the diagnosis. High-grade gliomas showed a higher frequency of PPM1D expression than low-grade gliomas (70.2% vs 16.7%, p < .001). In terms of molecular profiles, p53 expression showed a positive correlation with PPM1D expression (p < .001). There was a marginally negative relationship between PPM1D expression and IDH1 mutation (p = .062).
To assess the relationships between PPM1D expression, mRNA levels, and DNA copy-number alterations, RNAscope in situ hybridization, and FISH were conducted. Informative results were obtained in 77 cases. Thirty-five cases (45.5%, 35/77) were classified as demonstrating high PPM1D mRNA levels (Fig. 1B). There was a significant correlation between PPM1D protein expression and PPM1D mRNA expression (p = .035) (Table 2). Patients with PPM1D expression were found to have higher PPM1D mRNA expression than those without PPM1D expression (58.5%, 24/41 vs 31.4%, 11/35; p = .026). A large number of cases involving PPM1D positivity (90.2%, 37/41) showed mRNA expression (low or high expression), and over half of the cases (58.5%, 24/41) presented high PPM1D mRNA levels. The level of PPM1D positivity in the high PPM1D mRNA group was significantly higher than that in the low/no PPM1D mRNA group (68.6%, 24/35 vs 40.5%, 17/42; p = .026).
Of the cases examined using FISH analyses, 11 (14.3%) showed PPM1D gene amplification and four (5.2%) demonstrated relative copy gain (Fig. 1C). The majority of the cases with PPM1D gene amplification exhibited PPM1D positivity (81.8%, 9/11). The proportion of PPM1D positivity in the PPM1D amplification group was higher than that in the PPM1D nonamplification/relative copy gained group with borderline significance (81.8%, 9/11 vs 48.5%, 32/66; p = 0.053). However, there was a marginal correlation between PPM1D expression and PPM1D gene amplification (p = .079).
Age, diagnosis, IDH1 mutation, and PPM1D expression were significant predictors of survival in univariate Cox proportional hazard regression analyses (Table 3). The patients aged over 60 years presented an increased risk of poor prognosis (hazard ratio [HR] 3.97; 95% confidence interval [CI], 1.86 to 8.46; p < .001). Those diagnosed with glioblastoma, IDH-wildtype showed the worst prognosis (HR, 8.28; 95% CI, 1.71 to 40.01; p = .008). PPM1D expression was significantly associated with decreased survival (HR, 3.35; 95% CI, 1.42 to 7.90; p = .005), and survival in PPM1D-positive patients was significantly shorter than in PPM1D-negative patients (median overall survival [OS], 21 months [95% CI, 19 to not reached] vs not reached; p = .003) (Fig. 2A). Mutant IDH1 was a significant protective factor when compared with wild-type IDH1 (HR, 0.14; 95% CI, 0.05 to 0.38; p < .001), and the patients with mutant IDH1 showed significantly longer OS from the initial diagnosis (median OS, 102 months [95% CI, 102 to not reached] vs 19 months [95% CI, 14 to not reached]; p < .001) (Fig. 2B). In multivariate analyses, age (> 60 years) (HR, 2.55; 95% CI, 1.17 to 5.55; p = .018), PPM1D positivity (HR, 2.58; 95% CI, 1.08 to 6.17; p = .032), and IDH1 mutation (HR, 0.18; 95% CI, 0.06 to 0.53; p = .002) were independent prognostic factors (Table 3). Thus, the prognostic significance of PPM1D protein expression was further analyzed. Patients with mutant IDH1 and PPM1D-negative (IDH1mut/PPM1D-negative) tumors had the best OS, whereas patients with wild-type IDH1 and PPM1D expression (IDH1wt/PPM1D-positive) showed the worst OS (p < .001) (Fig. 2C).
Histologic diagnosis and grading were the gold standard for diagnosing gliomas. Recently, the well-established molecular markers, IDH and 1p/19q, were incorporated in the 2016 WHO classification [7]. In the present study, we explored the effect of PPM1D expression and its association with other molecular markers, including IDH and p53. To our knowledge, this is the first study regarding PPM1D expression in supratentorial diffuse astrocytic and oligodendroglial tumors. Patients with PPM1D-positive tumors had significantly worse OS than those without PPM1D-expressing tumors (p = .003) (Fig. 2A); in terms of prognostic utility, PPM1D expression was found to be comparable to IDH mutation status.
Genetic alterations of PPM1D are observed in various tumors, and are significantly associated with poor prognosis [23-29]. Our data showed that overexpression of p53 and that of PPM1D were positively correlated (p < .001) (Table 1). The results were in contrast to our expectation because p53 mutation and PPM1D overexpression were exclusively detected in tumors. p53 mutation status rather than p53 protein expression is clearly associated with poor prognosis in malignant gliomas [35,36]. In our study, PPM1D positivity was significantly correlated with its mRNA transcript levels (p = .035) and marginally associated with PPM1D gene amplification (p = .079). The degree of PPM1D-positivity in the PPM1D amplification group was greater than that in the PPM1D non-amplification/relative copy gained group (81.8% vs 48.5%, p = .053) (Table 2). Based on the data, we believe that PPM1D protein expression (55%, 46/84) may be increased by mRNA upregulation and DNA amplification of the PPM1D gene rather than functional wild-type p53 in this cohort. Recently, Zhang et al. [32] reported that p53 (58%, 19/33) and PPM1D (18%, 6/33) mutations demonstrate a mutually exclusive pattern in brainstem gliomas; moreover, OS rates are similar in patients with p53-mutated gliomas and those with PPM1D-mutated gliomas. The genomic landscape of supratentorial diffuse astrocytic and oligodendroglial tumors could be different from that of brainstem gliomas, which have two main driver mutations, p53 and PPM1D, in the absence of PPM1D overexpression. PPM1D mutation is very rare in gliomas arising outside the brainstem [32,37]. Our data suggest that PPM1D expression in supratentorial diffuse astrocytic and oligodendroglial tumors may have similar effects as PPM1D mutations in brainstem gliomas, resulting in stabilized and extended phosphatase activities of PPM1D.
In our study, multivariate analysis for OS according to various clinical and molecular parameters revealed that PPM1D is a significant molecular marker (HR, 2.58; p = .032) together with IDH1 mutation (HR, 0.18; p = .002). In accordance with previous studies [38,39], our data showed that p53 overexpression did not have prognostic effects, per univariate (p=.064) and multivariate (p = .986) analyses. The results suggest that p53 expression did not enhance the negative prognostic effect in terms of survival, although we observed a positive correlation between PPM1D and p53 expressions. In addition, we analyzed mRNA expression and DNA alteration in the PPM1D gene in 516 lower grade gliomas including grades II and III available for the PPM1D gene, using the cBioPortal for Cancer Genomics from TCGA data (Supplementary Table S1) [40,41]. Of the 40 cases with alterations, gene amplifications were found in 10 cases, deep deletion in 1, mRNA upregulation in 31, and mRNA downregulation in 2 were found (Supplementary Table S2, Supplementary Fig. S1A). The patients with up-regulated PPM1D transcripts and/or DNA amplification had a shorter OS than those who did not demonstrate these alterations (p = .031) (Supplementary Fig. S1B). The results further confirm that PPM1D expression could be a novel prognostic marker in supratentorial diffuse astrocytic and oligodendroglial tumors. We further characterized two prognostic factors: IDH1 mutation and PPM1D expression (Fig. 2C). Patients with IDH1mut/PPM1D-negative tumors had the most favorable OS; those with IDH1wt/PPM1D-positive tumors had the worst OS (p < .001). Considering the median OS for PPM1D-positive patients (21 months; 95% CI, 19 to not reached) and IDH1wt patients (19 months; 95% CI, 14 to not reached), these results suggest that PPM1D expression and wild-type IDH1 status seem to have an additive negative prognostic effect on survival in supratentorial gliomas and may promote tumorigenesis via different mechanisms.
In conclusion, our results indicate that PPM1D has the potential to be a molecular prognostic marker of adult diffuse astrocytic and oligodendroglial tumors and its prediction abilities are independent of the IDH1 and 1p19q co-deletion. Although these results need to be further validated, we hope to provide a basis for classifying this PPM1D-positive subset of gliomas and open up new opportunities for the treatment of such patients using PPM1D inhibitors.
Electronic Supplementary Material
Supplementary materials are available at Journal of Pathology and Translational Medicine (http://jpatholtm.org).
ACKNOWLEDGMENTS
This work was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), Ministry of Health, Welfare and Family Affairs, Republic of Korea (HI15C2111).
REFERENCES
3. Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013; 155:462–77.
4. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008; 455:1061–8.
5. Cancer Genome Atlas Research Network, Brat DJ, Verhaak RG, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015; 372:2481–98.

6. Chen R, Smith-Cohn M, Cohen AL, Colman H. Glioma subclassifications and their clinical significance. Neurotherapeutics. 2017; 14:284–97.

7. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016; 131:803–20.

8. Reuss DE, Sahm F, Schrimpf D, et al. ATRX and IDH1-R132H immunohistochemistry with subsequent copy number analysis and IDH sequencing as a basis for an "integrated" diagnostic approach for adult astrocytoma, oligodendroglioma and glioblastoma. Acta Neuropathol. 2015; 129:133–46.

9. Suzuki H, Aoki K, Chiba K, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015; 47:458–68.

10. Lass U, Numann A, von Eckardstein K, et al. Clonal analysis in recurrent astrocytic, oligoastrocytic and oligodendroglial tumors implicates IDH1-mutation as common tumor initiating event. PLoS One. 2012; 7:e41298.
11. Labussière M, Idbaih A, Wang XW, et al. All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology. 2010; 74:1886–90.

12. Nagpal J, Jamoona A, Gulati ND, et al. Revisiting the role of p53 in primary and secondary glioblastomas. Anticancer Res. 2006; 26:4633–9.
13. Tabouret E, Nguyen AT, Dehais C, et al. Prognostic impact of the 2016 WHO classification of diffuse gliomas in the French POLA cohort. Acta Neuropathol. 2016; 132:625–34.

14. Dudgeon C, Shreeram S, Tanoue K, et al. Genetic variants and mutations of PPM1D control the response to DNA damage. Cell Cycle. 2013; 12:2656–64.
15. Lee DH, Chowdhury D. What goes on must come off: phosphatases gate-crash the DNA damage response. Trends Biochem Sci. 2011; 36:569–77.

16. Le Guezennec X, Bulavin DV. WIP1 phosphatase at the crossroads of cancer and aging. Trends Biochem Sci. 2010; 35:109–14.

17. Lu X, Nguyen TA, Moon SH, Darlington Y, Sommer M, Donehower LA. The type 2C phosphatase Wip1: an oncogenic regulator of tumor suppressor and DNA damage response pathways. Cancer Metastasis Rev. 2008; 27:123–35.

18. Song JY, Ryu SH, Cho YM, et al. Wip1 suppresses apoptotic cell death through direct dephosphorylation of BAX in response to gamma-radiation. Cell Death Dis. 2013; 4:e744.
19. Bulavin DV, Demidov ON, Saito S, et al. Amplification of PPM1D in human tumors abrogates p53 tumor-suppressor activity. Nat Genet. 2002; 31:210–5.

20. Kleiblova P, Shaltiel IA, Benada J, et al. Gain-of-function mutations of PPM1D/Wip1 impair the p53-dependent G1 checkpoint. J Cell Biol. 2013; 201:511–21.

21. Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev. 2005; 19:1162–74.

22. Lu X, Ma O, Nguyen TA, Jones SN, Oren M, Donehower LA. The Wip1 phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell. 2007; 12:342–54.

23. Hirasawa A, Saito-Ohara F, Inoue J, et al. Association of 17q21-q24 gain in ovarian clear cell adenocarcinomas with poor prognosis and identification of PPM1D and APPBP2 as likely amplification targets. Clin Cancer Res. 2003; 9:1995–2004.
24. Hu W, Feng Z, Modica I, et al. Gene amplifications in well-differentiated pancreatic neuroendocrine tumors inactivate the p53 pathway. Genes Cancer. 2010; 1:360–8.

25. Lambros MB, Natrajan R, Geyer FC, et al. PPM1D gene amplification and overexpression in breast cancer: a qRT-PCR and chromogenic in situ hybridization study. Mod Pathol. 2010; 23:1334–45.

26. Loukopoulos P, Shibata T, Katoh H, et al. Genome-wide array-based comparative genomic hybridization analysis of pancreatic adenocarcinoma: identification of genetic indicators that predict patient outcome. Cancer Sci. 2007; 98:392–400.

27. Saito-Ohara F, Imoto I, Inoue J, et al. PPM1D is a potential target for 17q gain in neuroblastoma. Cancer Res. 2003; 63:1876–83.
28. Tan DS, Lambros MB, Rayter S, et al. PPM1D is a potential therapeutic target in ovarian clear cell carcinomas. Clin Cancer Res. 2009; 15:2269–80.

29. Yu E, Ahn YS, Jang SJ, et al. Overexpression of the wip1 gene abrogates the p38 MAPK/p53/Wip1 pathway and silences p16 expression in human breast cancers. Breast Cancer Res Treat. 2007; 101:269–78.

30. Liang C, Guo E, Lu S, et al. Over-expression of wild-type p53-induced phosphatase 1 confers poor prognosis of patients with gliomas. Brain Res. 2012; 1444:65–75.

31. Wang P, Rao J, Yang H, Zhao H, Yang L. Wip1 over-expression correlated with TP53/p14(ARF) pathway disruption in human astrocytomas. J Surg Oncol. 2011; 104:679–84.

32. Zhang L, Chen LH, Wan H, et al. Exome sequencing identifies somatic gain-of-function PPM1D mutations in brainstem gliomas. Nat Genet. 2014; 46:726–30.

33. Chuman Y, Kurihashi W, Mizukami Y, Nashimoto T, Yagi H, Sakaguchi K. PPM1D430, a novel alternative splicing variant of the human PPM1D, can dephosphorylate p53 and exhibits specific tissue expression. J Biochem. 2009; 145:1–12.

34. Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004; 10:7252–9.
35. Wang X, Chen JX, Liu JP, You C, Liu YH, Mao Q. Gain of function of mutant TP53 in glioblastoma: prognosis and response to temozolomide. Ann Surg Oncol. 2014; 21:1337–44.

36. Watanabe T, Katayama Y, Yoshino A, Komine C, Yokoyama T. Deregulation of the TP53/p14ARF tumor suppressor pathway in low-grade diffuse astrocytomas and its influence on clinical course. Clin Cancer Res. 2003; 9:4884–90.
37. Jones C, Baker SJ. Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nat Rev Cancer. 2014; 14:651–61.

38. Karsy M, Neil JA, Guan J, Mahan MA, Colman H, Jensen RL. A practical review of prognostic correlations of molecular biomarkers in glioblastoma. Neurosurg Focus. 2015; 38:E4.

40. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–4.
Fig. 1.
A representative case of high-grade glioma. (A) Protein phosphatase magnesium-dependent 1δ (PPM1D) staining shows a high percentage of cells with positive protein expression in both the nucleus and cytoplasm in the anaplastic oligodendroglioma. (B) RNAscope in situ hybridization analysis shows positive expression of PPM1D shown as red dots. (C) Fluorescence in situ hybridization shows amplified PPM1D gene copies as red spots.
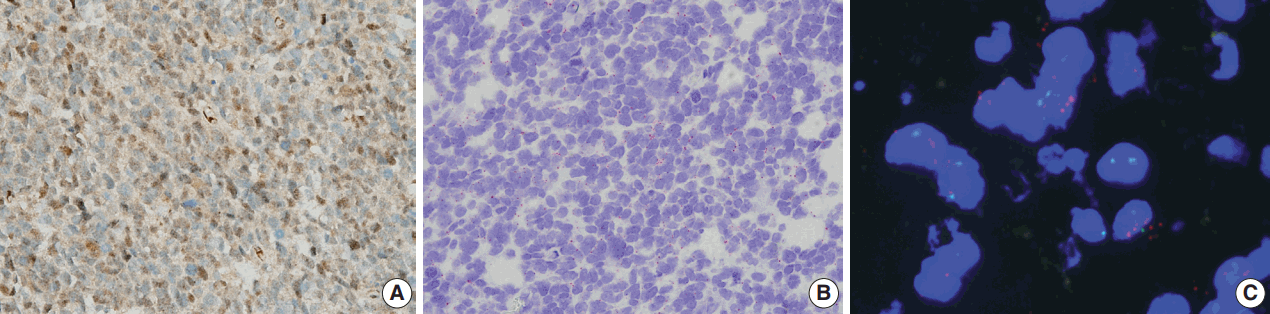
Fig. 2.
Kaplan-Meier survival curves of patients with supratentorial diffuse astrocytic and oligodendroglial tumors. (A) Comparison of patients with protein phosphatase magnesium-dependent 1δ (PPM1D) expression and those without PPM1D expression. (B) Comparison of patients with isocitrate dehydrogenase 1 (IDH1) mutation versus those with wild-type IDH1. (C) Comparison of patients with PPM1D expression versus those without PPM1D expression with or without an additional IDH1 mutation. Log-rank tests for a, b, and c yielded p=.003, p<.001, and p< .001, respectively.
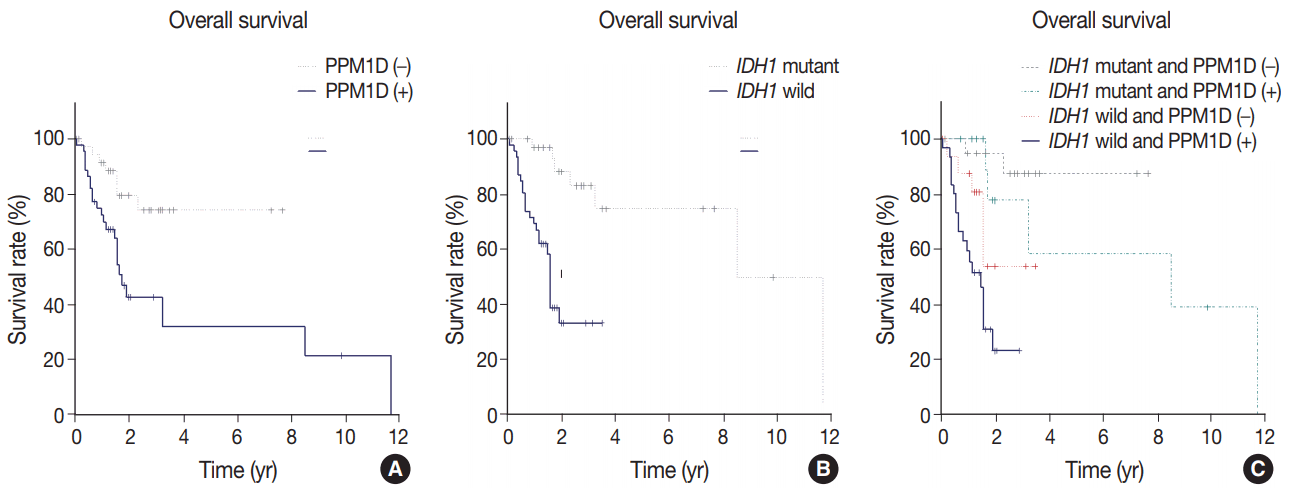
Table 1.
Baseline characteristics of the enrolled patients
Table 2.
Correlation between PPM1D protein expression and genetic variables of PPM1D
Table 3.
Cox proportional hazard regression analysis for overall survival of the patients with supratentorial glioma




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download