This article has been
cited by other articles in ScienceCentral.
Abstract
Background
Mixed gliomas, such as oligoastrocytomas (OA), anaplastic oligoastrocytomas, and glioblastomas (GBMs) with an oligodendroglial component (GBMO) are defined as tumors composed of a mixture of two distinct neoplastic cell types, astrocytic and oligodendroglial. Recently, mutations ATRX and TP53, and codeletion of 1p/19q are shown to be genetic hallmarks of astrocytic and oligodendroglial tumors, respectively. Subsequent molecular analyses of mixed gliomas preferred the reclassification to either oligodendroglioma or astrocytoma. This study was designed to apply genetically integrated diagnostic criteria to mixed gliomas and determine usefulness and prognostic value of new classification in Korean patients.
Methods
Fifty-eight cases of mixed OAs and GBMOs were retrieved from the pathology archives of Seoul National University Hospital from 2004 to 2015. Reclassification was performed according to genetic and immunohistochemical properties. Clinicopathological characteristics of each subgroup were evaluated. Overall survival was assessed and compared between subgroups.
Results
We could reclassify all mixed OAs and GBMOs into either astrocytic or oligodendroglial tumors. Notably, 29 GBMOs could be reclassified into 11 cases of GBM, IDH-mutant, 16 cases of GBM, IDH-wildtype, and two cases of anaplastic oligodendroglioma, IDH mutant. Overall survival was significantly different among these new groups (p<.001). Overall survival and progression-free survival were statistically better in gliomas with IDH mutation, ATRX mutation, no microscopic necrosis, and young patient age (cut off, 45 years old).
Conclusions
Our results strongly suggest that a genetically integrated diagnosis of glioma better reflects prognosis than former morphology-based methods.
Keywords: Oligoastrocytoma, Glioblastoma with oligodendroglioma component, WHO classification, Genetics, Integrated diagnosis
Mixed gliomas, such as oligoastrocytomas (OA), anaplastic oligoastrocytomas (AOA), and glioblastomas (GBMs) with oligodendroglial component (GBMO), are defined as tumors composed of a mixture of two distinct neoplastic cell types; astrocytic and oligodendroglial. However, there have been controversies as to whether these mixed tumors are really distinct entities. First, research has found no significant difference in survival between mixed gliomas versus either astrocytic or oligodendroglial tumors, such as GBMO versus GBM [
1], anaplastic astrocytoma (AA) versus AOA [
2], anaplastic oligodendroglioma (AO) versus AOA [
3], diffuse astrocytoma (DA) versus OA [
4], and oligodendroglioma versus OA [
4]. Second, results of genetic studies suggest that morphologically mixed OA or ambiguous glioma is genetically monoclonal and similar to either DA or oligodendroglioma, rather than a mixture of them [
5]. In addition, it is well known that the diagnosis of mixed glioma shows a high interobserver variability [
6].
The accumulation of research on gliomas has led to a growth in understanding of its molecular aspects. Previous molecular study of astrocytic tumors revealed frequent mutations in
IDH, ATRX, and
TP53, suggesting that these mutations might be genetic features of astrocytic tumors [
7,
8]. On the other hand, concurrent deletion of chromosomal arms 1p and 19q constitute a hallmark alteration in oligodendroglial tumors [
9]. Because 1p/19q codeleted gliomas are strongly associated with
IDH mutation [
10],
IDH mutation is considered as a genetic feature of oligodendroglioma. The disclosure of these molecular aberrations of glioma led to the discussion on whether to include these molecular markers in the 2016 revised World Health Organization (WHO) classification of central nervous system tumors. As such, neuropathologists with expertise in molecular diagnosis met in a meeting held in Haarlem [
11]. This “International Society of Neuropathology (ISN)-Haarlem Consensus Meeting” decided to include the ambiguous category of “DA, 1p/19q-codeleted”; however, this class turned out to be oligodendroglioma with an astrocytic phenotype. In August 2014, Sahm
et al. [
12] suggested no longer using the term “OA” because
in situ molecular genetics showed that previously diagnosed OAs were largely divided into oligodendroglioma with reactive astrogliosis or astrocytoma with partial oligodendroglial morphology with no genetic basis of the oligodendroglial component. Rather, the latter showed genetic aberration compatible with DA in the whole tumor area [
12]. Subsequently, a genetically integrated diagnostic approach was proposed [
13]. With respect to both overall survival (OS) and time until treatment failure, the integrated approach separated the diagnostic groups more stringently, and it also proved to be more successful in the outcome prediction [
13]. Not only in terms of prognosis, but also for predictive information, the previous studies suggest that
IDH, ATRX, and 1p/19q status can offer useful criteria and aid in selection of proper therapy for the patients [
14,
15].
With this background, we reclassified the archives of mixed gliomas at Seoul National University Hospital, including GBMO, by the new genetically integrated diagnostic criteria. This study was designed to determine the usefulness of a genetically integrated diagnostic approach and the prognostic power of a new classification system in Korean patients.
MATERIALS AND METHODS
Study population
Formalin-fixed paraffin-embedded (FFPE) tissues from a total of 58 patients were obtained from the archives of the Department of Pathology, Seoul National University Hospital. All patients were Korean (
Table 1). Age at first diagnosis was between 14 and 73 years old (median: previously OA, 36 years; previously GBMO, 56 years). All cases were diagnosed between 2004 and 2015. Selected cases had distinct morphological features of mixed astrocytic and oligodendroglial component or mixed GBM and oligodendroglial component. We used a 10% cut-off for the minimum area of oligodendroglial or astrocytic component. Tumors were graded according to the 2007 WHO guidelines. The series contained 15 OAs, 14 AOAs, and 29 GBMOs (
Fig. 1). Histopathologic slides were reviewed by two pathologists (S.H. Park, S.I. Kim). Immunohistochemical stainings for IDH1 (clone H09, Dianova, Hamburg, Germany) and ATRX (Sigma-Aldrich, St. Louis, MO, USA) were performed in all 58 cases. If IDH1 immunohistochemistry (IHC) was negative, direct sequencing of
IDH1/IDH2 was performed. Fluorescent
in situ hybridization (FISH) using Vysis probes (Abbott, Des Plaines, IL, USA) to detect 1p/19q status was done in all 58 cases. Survival plots according to the clinical, histological, and molecular characteristics of tumors were analyzed using Kaplan-Meier (K-M) survival analysis.
With the results of our genetic studies, we reclassified tumors based on the diagram by Reuss
et al. [
13]. If the ATRX IHC was negative, it was classified as astrocytic, independent of its IDH status. Tumors were then subdivided by IDH status based on IHC and direct sequencing. In contrast, all tumors with nuclear ATRX expression were divided by 1p/19q status using FISH. If 1p/19q were non-codeleted, tumors were astrocytic tumors. Then, the tumors were also subdivided by IDH status. If codeletion of 1p/19q and
IDH mutation were present, they were defined as oligodendroglial tumors. Next, we graded the tumors according to the new 2016 WHO guidelines. Lastly, 58 cases were reclassified into seven distinct subgroups: 1) DA, IDH-mutant (DA IDH-m), 2) DA, IDH-wildtype (DA IDH-w), 3) AA, IDH-mutant (AA IDH-m), 4) AA, IDH-wildtype (AA IDH-w), 5) AO, IDH-mutant (AO IDH-m), 6) GBM, IDH-mutant (GBM IDH-m), and 7) GBM, IDH-wildtype (GBM IDH-w). We performed further classification of GBM into primary and secondary GBM. Secondary GBM was defined as cases with previous pathology-proven lower grade tumors.Then, we compared the survival plots between reclassified entities using K-M survival analysis.
This study abided by the world Medical Association Declaration of Helsinki recommendation and was approved by the Institutional Review Board of Seoul National University Hospital (IRB No. 1508-004-690). Written informed consents were obtained.
Hematoxylin and eosin staining and IHC
FFPE (10% neutral buffered formalin, routinely processed, and paraffin-embedded) tissue sections (2–3 μm thick) were cut for hematoxylin and eosin staining and IHC. Tissue sections were stained with anti-IDH1 R132H (H09) monoclonal antibody (Dianova) using a 1:100 dilution, anti-ATRX polyclonal antibody HPA001906 (Sigma-Aldrich) using a 1:300 dilution, and anti-p53 monoclonal antibody, DO-7 code M7001 (Dako, Glostrup, Denmark) using a 1:1000 dilution. IHC staining was carried out using a standard avidin–biotin peroxidase method. Tumors were interpreted as positive for p53 expression if ≥ 20% of neoplastic cells showed distinct nuclear staining.
DNA extraction
Tumor areas were manually micro-dissected using 6-μm unstained tissue sections made from FFPE tissue. DNA was isolated from the micro-dissected tissue using a DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions.
Polymerase chain reaction amplification and sequencing of IDH1 and IDH2
Template DNA (1 μL) was added to 100 μL of polymerase chain reaction (PCR) solution (10 μL of 10 × magnesium Taq High-Fidelity [HF] buffer, 10 μL deoxynucleotide triphosphate [dNTP] mixture with 2 mM magnesium, 5 μL of 10 pmol primer [2 ×], 1 μL of magnesium Taq-HF polymerase, and distilled water). The IDH1-Forward (F)/IDH1-Reverse (R) and IDH2-F/IDH2-R primers (
Table 2) were used with the following program: 35 cycles of 95℃ for 30 seconds, 55℃ for 30 seconds, and 72℃ for 60 seconds. The product sizes were 130-base pair (bp) (IDH1) and 293-bp (IDH2). Unincorporated PCR primers and dNTPs were removed from the PCR products using a Montage PCR Clean-up Kit (Millipore, Billerica, MA, USA).
Purified products were sequenced using the same primers. Sequencing was performed using a BigDye Terminator Cycle Sequencing Kit v. 3.1 (Applied Biosystems, Foster City, CA, USA). Sequencing products were resolved on an automated DNA sequencing system, model 3730XL (Applied Biosystems).
Fluorescence in situ hybridization
FISH with Vysis probes was used to assess 1p/19q status. Sections (3 μm thick) were first deparaffinized in xylene, incubated with 0.3% pepsin in 10 mM HCl at 37℃ for 10 minutes, and boiled with citrate buffer (pH 6.0) in a microwave. Sections were then incubated in 1 M NaSCN for 35 minutes at 80℃, immersed in the pepsin solution, and fixed in 10% neutral buffered formalin. 1p36/1q25 and 19q13/19p13 labeled locusspecific dual-color probes (Abbott Molecular) were used according to the manufacturer’s protocol. We applied the probe mixture to the slides and incubated them in a humidified atmosphere with HYBrite (Abbott Molecular) at 73℃ for 5 minutes for simultaneous denaturation of the probe and target DNA. Then, we cooled the samples to 37℃ and incubated for 19 hours to hybridize the probe and target DNA. The slides were submerged in 0.4× SSC buffer/0.3% NP-40 for 2 minutes at room temperature, followed by incubation in 2× SSC/0.1% NP-40 for 5 minutes at 73℃.
Statistical analysis
OS and progression-free survival (PFS) were estimated using K-M survival analysis and were compared using the log-rank test. All statistical analyses were performed with SPSS ver. 22 (IBM Corp., Armonk, NY, USA); p < .05 was considered significant.
RESULTS
Clinicopathological information of patients is provided in
Table 1. Our reclassification results are summarized in
Table 3 and
Fig. 2. Our reclassification analysis showed that the largest fraction of mixed OA was AA IDH-m (50%). The largest part of mixed AOA was also AA IDH-m (42.9%), followed by AA IDH-w (35.7%) and AO (21%). The largest fraction of GBMO was conventional GBM IDH-w (55.2%), followed by GBM IDH-m (37.9%) and only 6.9% AO. Except for one undetermined case of GBM IDH-w, 14 out of 16 GBM IDH-w cases (87.5%) were primary GBM and one case (6.3%) was secondary GBM. In 11 cases of GBM IDH-m, seven cases (63.6%) were primary GBM and four cases (36.4%) were secondary GBM. Therefore, the incidence of primary GBMs (21/27, 77.8%) derived from those previously diagnosed as mixed gliomas was less than the well-known incidence of primary GBM (90%–95%).
When we carried out K-M survival analysis, two cases of DA IDH-w were excluded; one patient died of suddenly developed progressive hydrocephalus of an unknown cause and the other patient didn’t show up. One case of AO IDH-m was also excluded because the patient died of a worsened general condition from multiple diseases, including pulmonary thromboembolism with basal ganglia and temporal lobe infarction during hospitalization. Recent studies report a very similar incidence of stroke between cancer and non-cancer patients; therefore, it is reasonable to think that stroke was an independent variable for survival in this case, whether the tumor was a causative factor of stroke or not [
16].
OS in different clinical, histological, and molecular subgroups has been evaluated (
Fig. 3). Prognosis was better in the younger (age < 45 years) (p = .010) and IDH-mutant (p =. 007) subgroups. However, prognosis was worse in subgroups with microscopic necrosis (p < .001) and ATRX-positive IHC (p = .031). Similar results were drawn with PFS (
Fig. 4). Other characteristics failed to show statistical significance. The subgroup with the 1p/19q co-deletion had a tendency of better prognosis in terms of OS (p = .067) and PFS (p = .077), despite the lack of statistical significance (
Figs. 3F,
4F). Of those compared, all 1p/19q codeleted patients fully survived the study completion. Also, those with p53 overexpression (positive in > 20%) had a tendency for worse prognosis in terms of OS (p = .054) and PFS (p = .208) (
Figs. 3E,
4E).
All these results with demographic characteristics are summarized in
Table 4.
After re-classification of mixed gliomas, we compared survival plots between the reclassified entities using K-M survival analysis. In terms of OS as well as PFS, the genetically integrated diagnostic approach showed clear statistical significance (p < .001) (
Fig. 5).
DISCUSSION
Mixed gliomas have been previously defined as diffusely infiltrating gliomas composed of a mixture of two distinct neoplastic cell types morphologically resembling the tumor cells in oligodendroglial or astrocytic tumors [
9]. However, inter-institutional and inter-observer variations of mixed gliomas have resulted in low diagnostic reproducibility and reliability.
The ISN-Haarlem’s recommendations for a genetically integrated diagnosis for the classification of diffuse gliomas suggested the removal of mixed glioma entity [
11]. We reclassified mixed OA (including GBMO) into either astrocytic or oligodendroglial tumors in agreement with the emphasis by Sahm
et al. [
12].
Here, we applied the new criteria to the 58 cases that were previously diagnosed as mixed gliomas. The purpose of our study was to determine the usefulness of genetically integrated classification, true incidence of diffuse gliomas, and prognostic value of a new classification system. Above all, we sought to find out whether there are any intermediate cases or genuine mixed gliomas.
We primarily used Reuss
et al.’s diagnostic criteria [
13]; therefore, ATRX IHC and 1p/19q FISH were used to determine whether the tumor was of astrocytic or oligodendroglial lineage.
Among DA and AA tumors, ATRX-negativity and -positivity were found in 88% and 12%, respectively. Six cases which showed 1p/19q codeletion were also ATRX-positive and IDH1-mutated; therefore, the diagnosis of AO was certain. Among our 27 GBMs, ATRX-positive and -negative cases were 55.6% and 44.4%, respectively. Among ATRX-negative GBMs, 50% (6/12) were secondary, 41.7% (5/12) were primary, and 8.3% (1/12) were unconfirmed. Thus, half of ATRX-negative GBMs were secondary GBM, but among primary GBMs, 27.8% were ATRX-negative.
Using these criteria, our 58 cases could be classified as either astrocytic or oligodendroglial. None showed mixed genetic features of astrocytic and oligodendroglial. Our results also indicate that the codeletion status of 1p/19q is the most important criteria in the classification of diffuse gliomas into astrocytic or oligodendroglial, and could serve as a useful biomarker in the future diagnosis.
We evaluated all clinical, histological, and molecular characteristics using K-M survival analysis. OS and PFS were better in gliomas with
IDH mutation,
ATRX mutation, absence of microscopic necrosis, and young patient age (cut off, 45 years old). Although the impact of
ATRX mutation on PFS had no statistical significance with log-rank test (p = .073), the Wilcoxon test and Tarone-Ware test showed statistical significance (p = .010 and p = .016, respectively). This confirms the results of previous studies on the impact of
IDH and
ATRX on prognosis [
17,
18]. Because most tumors with microscopic necrosis were diagnosed as either grade IV GBM IDH-m or GBM IDH-w, it is obvious that tumors with no necrosis showed better prognosis. Regarding the patient age, this study also confirms the results of a previous study [
1]. Increase in age is associated with more high-grade glial tumors or poorer general conditions; therefore, elderly people could have a poorer prognosis [
9]. Among patients with tumors of the same grade, one of the most favorable prognostic factors is young age (cut off, 45 years old) [
9]. Similar results can be observed in oligodendroglial tumors [
19].
In this study, 1p/19q status did not achieve statistical significance on OS or PFS. However, excluding one case, of which the cause of death was uncertain, all 1p/19q codeleted patients fully survived the follow-up period. Therefore, the lack of statistical significance might be derived from the short term follow-up period or a small number of patients.
With regard to both OS and PFS, the genetically integrated diagnostic approach showed statistically significant subgroups, which confirm the results of previous studies carried out by Reuss
et al. [
13]. Our reclassification revealed that some patients should have received different treatment based on our genetically integrated diagnosis. Two patients initially diagnosed with GBMO were re-diagnosed with AO. One had primarily concurrent chemoradiation therapy and the other had primarily chemotherapy alone. However, according to our hospital protocol, the primary choice for AO is craniotomy and tumor removal with or without adjuvant therapy. Although reclassification itself might produce change in the treatment protocol of gliomas, it can also be useful in the selection of proper therapy for the patients.
ATRX negativity and p53 positivity directly indicated secondary GBM; however, among these tumors, four cases were primary GBM, which may indicate rapid progression from low grade tumors.
It is well known that astrocytic tumors have a genetic hallmark of
ATRX and
TP53 mutation [
7,
8]. In the genetically integrated diagnostic flow,
ATRX mutation proved more useful as
TP53 mutation was shown to be poorly detected by IHC [
20]. In cases with intact ATRX, a 1p/19q assay should be carried out for designation of oligodendroglial tumor. Since generally extensive p53 staining is regarded as indicative of astrocytoma, p53 IHC might also be helpful in differential diagnosis of those cases. Therefore, we tried to find for a proper cutoff value of p53 positivity for astrocytic designation in our study. The cut-off value of 20% divided the subgroup stringently. However, 20% cut off of p53 had no statistical significance both in OS (p = .054) and PFS (p = .208). In our study, astrocytic tumors had p53 positivity in an average of 42.1%, with a standard deviation of 32.31%. If astrocytic tumors were entirely negative for p53 IHC, those cases should be re-verified by alternative tools, such as next generation sequencing or restriction fragment length polymorphism for detecting loss of function-mutation or methylation of p53. Further studies on staining pattern or correlation with other clinicopathological factors might be helpful.
To compare prognosis, 55 patients with previously diagnosed mixed GBMO by 2007 WHO classification were classified into six groups, and there was a statistically significant difference (p = .000); however, the number of patients was limited. Thus, if longer follow-up or more prospective studies are ensured in more patients, some prognostic factors that were not statistically significant in current studies may turn out to be statistically significant.
In conclusion, we successfully reclassified previously diagnosed mixed gliomas into either astrocytic or oligodendroglial tumors with a genetically integrated diagnostic approach. Our study suggests that genetic information, including ATRX and IDH mutation and 1p/19q co-deletion, has powerful prognostic and predictive value. In addition, this information can aid in classifying brain tumors into more distinct categories. Therefore, the integration of phenotypes and genotypes is clearly necessary in the routine practice of brain tumor diagnoses.
ACKNOWLEDGMENTS
This study was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number : HI14C1277).
REFERENCES
1. Myung JK, Cho HJ, Kim H, et al. Prognosis of glioblastoma with oligodendroglioma component is associated with the IDH1 mutation and MGMT methylation status. Transl Oncol. 2014; 7:712–9.
2. Jiang H, Ren X, Cui X, et al. 1p/19q codeletion and IDH1/2 mutation identified a subtype of anaplastic oligoastrocytomas with prognosis as favorable as anaplastic oligodendrogliomas. Neuro Oncol. 2013; 15:775–82.
3. Tortosa A, Viñolas N, Villà S, et al. Prognostic implication of clinical, radiologic, and pathologic features in patients with anaplastic gliomas. Cancer. 2003; 97:1063–71.

4. Okamoto Y, Di Patre PL, Burkhard C, et al. Population-based study on incidence, survival rates, and genetic alterations of low-grade diffuse astrocytomas and oligodendrogliomas. Acta Neuropathol. 2004; 108:49–56.

5. Ohgaki H. Contribution of molecular biology to the classification of low-grade diffuse glioma. In: Duffau H, ed. Diffuse low-grade gliomas in adults. London: Springer;2013. p. 61–72.
6. Kros JM, Gorlia T, Kouwenhoven MC, et al. Panel review of anaplastic oligodendroglioma from European Organization For Research and Treatment of Cancer Trial 26951: assessment of consensus in diagnosis, influence of 1p/19q loss, and correlations with outcome. J Neuropathol Exp Neurol. 2007; 66:545–51.
7. Killela PJ, Pirozzi CJ, Reitman ZJ, et al. The genetic landscape of anaplastic astrocytoma. Oncotarget. 2014; 5:1452–7.

8. Liu XY, Gerges N, Korshunov A, et al. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol. 2012; 124:615–25.
9. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. WHO classification of tumours of the central nervous system. 4th ed. Lyon: IARC Press;2016.
10. Labussière M, Idbaih A, Wang XW, et al. All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology. 2010; 74:1886–90.
11. Louis DN, Perry A, Burger P, et al. International Society of Neuropathology: Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol. 2014; 24:429–35.
12. Sahm F, Reuss D, Koelsche C, et al. Farewell to oligoastrocytoma: in situ molecular genetics favor classification as either oligodendroglioma or astrocytoma. Acta Neuropathol. 2014; 128:551–9.
13. Reuss DE, Sahm F, Schrimpf D, et al. ATRX and IDH1-R132H immunohistochemistry with subsequent copy number analysis and IDH sequencing as a basis for an “integrated” diagnostic approach for adult astrocytoma, oligodendroglioma and glioblastoma. Acta Neuropathol. 2015; 129:133–46.

14. van den Bent MJ, Brandes AA, Taphoorn MJ, et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol. 2013; 31:344–50.
15. Houillier C, Wang X, Kaloshi G, et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in lowgrade gliomas. Neurology. 2010; 75:1560–6.
16. Grisold W, Oberndorfer S, Struhal W. Stroke and cancer: a review. Acta Neurol Scand. 2009; 119:1–16.

17. Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009; 360:765–73.
18. Haberler C, Wöhrer A. Clinical neuropathology practice news 2-2014: ATRX, a new candidate biomarker in gliomas. Clin Neuropathol. 2014; 33:108–11.

19. Love S, Perry A, Ironside J, Budka H. Greenfield's neuropathology. 9th ed. Boca Raton: CRC Press;2015.
20. Louis DN, von Deimling A, Chung RY, et al. Comparative study of p53 gene and protein alterations in human astrocytic tumors. J Neuropathol Exp Neurol. 1993; 52:31–8.

Fig. 1.
Hematoxylin and eosin features of a case that was previously diagnosed as glioblastoma with oligodendroglial component. This case turned out to be glioblastoma, IDH-wildtype in new classifications.
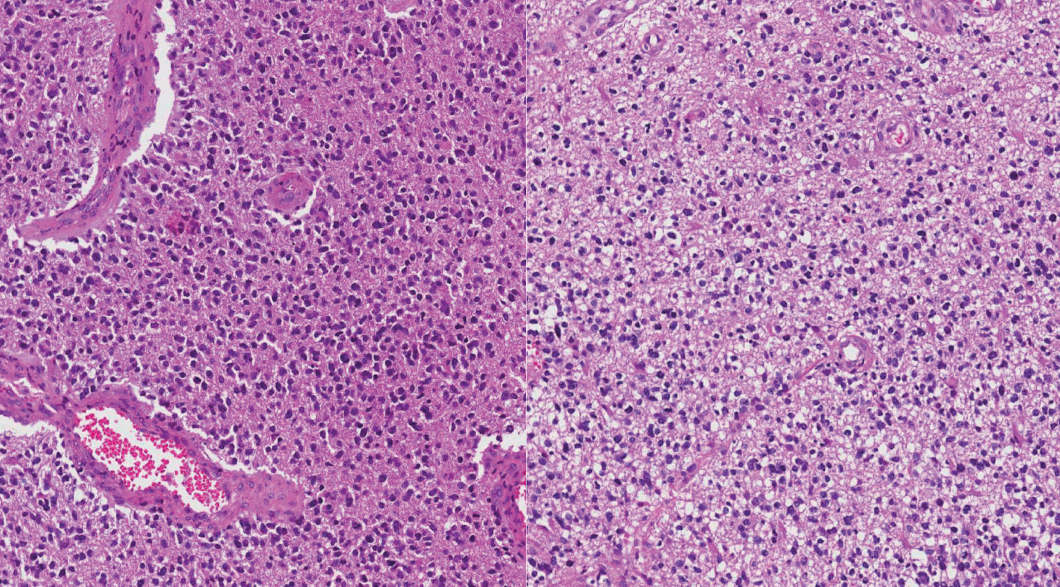
Fig. 2.
Change in diagnosis after applying molecular genetics integrated diagnostic criteria. OA, oligoastrocytoma; AOA, anaplastic oligoastrocytoma; GBMO, glioblastomas with an oligodendroglial component; DA, diffuse astrocytoma; m, mutant; w, wildtype; AA, anaplastic astrocytoma; AO, anaplastic oligodendroglioma; GBM, glioblastoma.
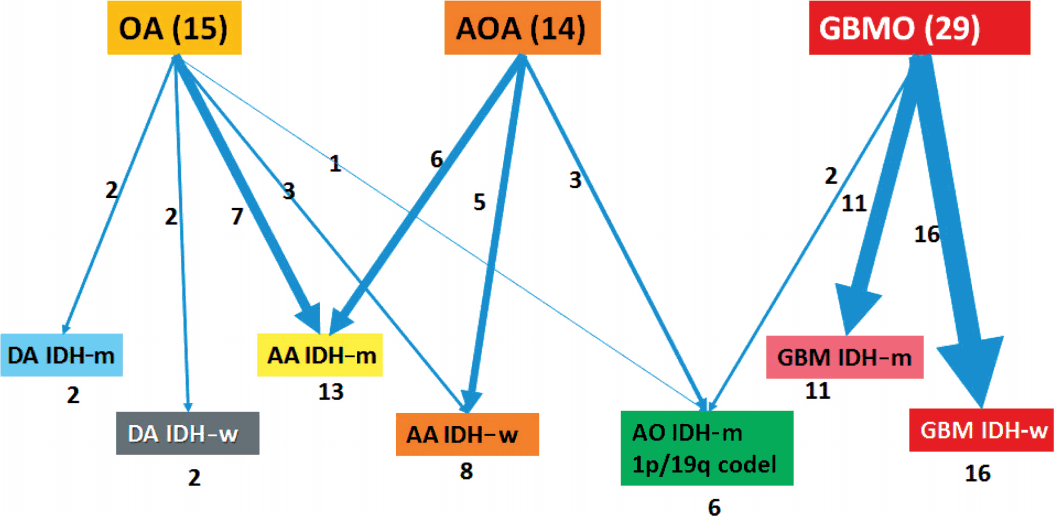
Fig. 3.
Overall survival (OS) in different clinical, histological, and molecular subgroups. (A) Age over 45 years. (B) Necrosis. (C) IDH mutation status. (D) ATRX methylation status. (E) p53 positivity (more than 20% of tumor in immunohistochemistry). (F) 1p/19q-codeletion status.
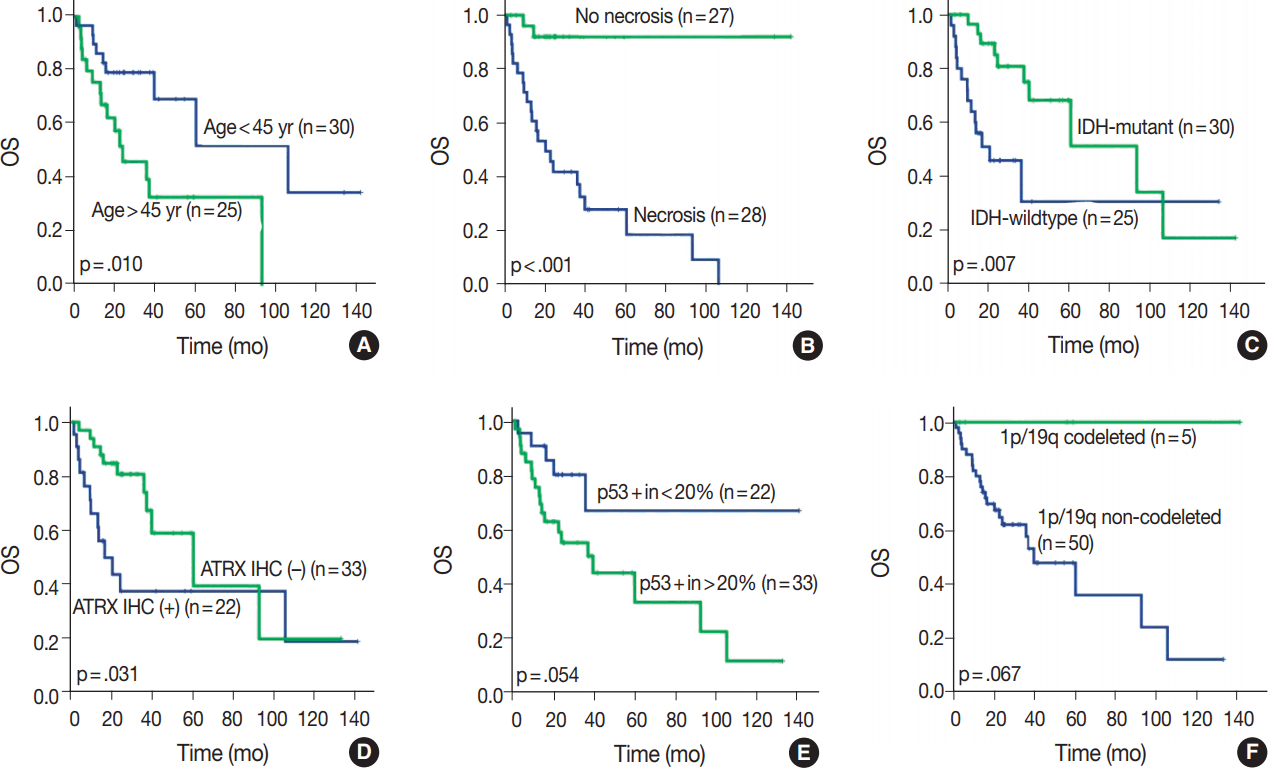
Fig. 4.
Progression-free survival (PFS) in different clinical, histological, and molecular subgroups. (A) Age over 45. (B) Necrosis. (C) IDH mutation status. (D) ATRX methylation status. (E) p53 positivity (more than 20% of tumor in IHC). (F) 1p/19q-codeletion status.
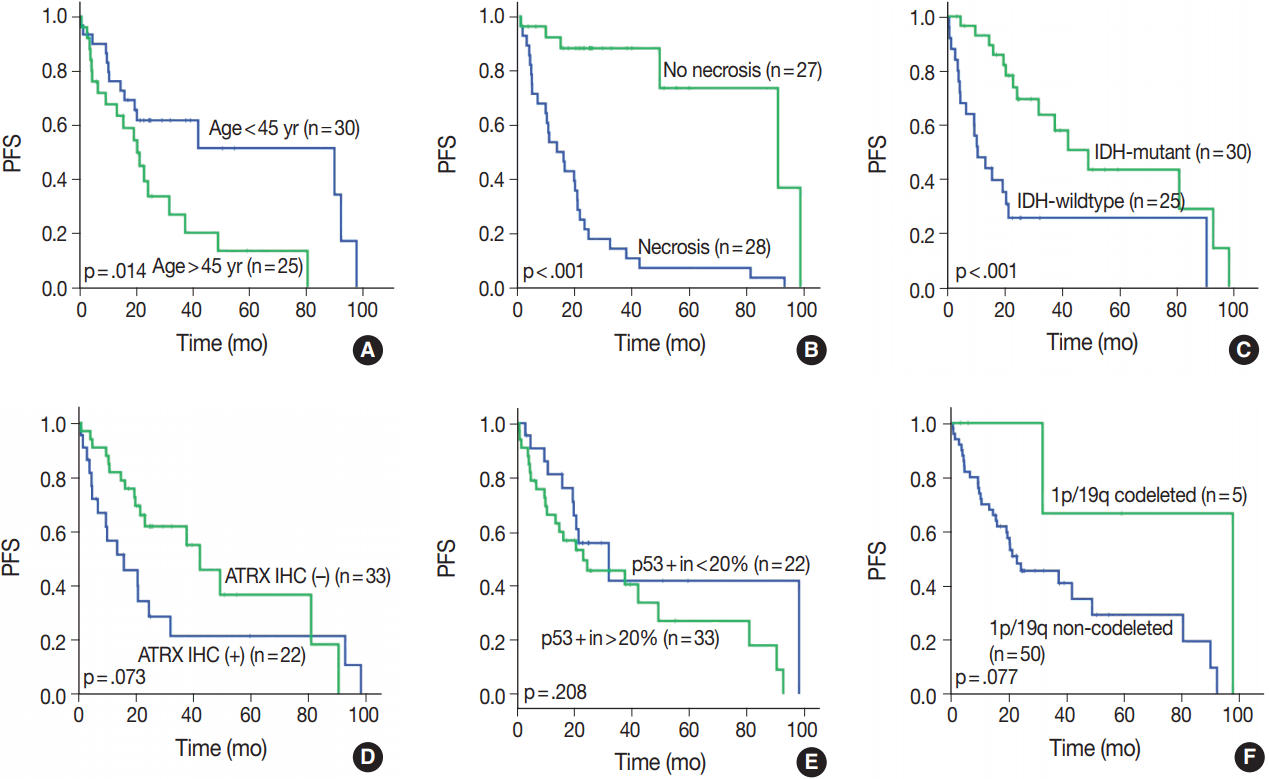
Fig. 5.
Overall survival (A) and progression-free survival (B) in rediagnosed subgroups. AA, anaplastic astrocytoma; DA, diffuse astrocytoma; AO, anaplastic oligodendroglioma; GBM, glioblastoma; IDH-m, IDH-mutant; IDH-w, IDH-wildtype.
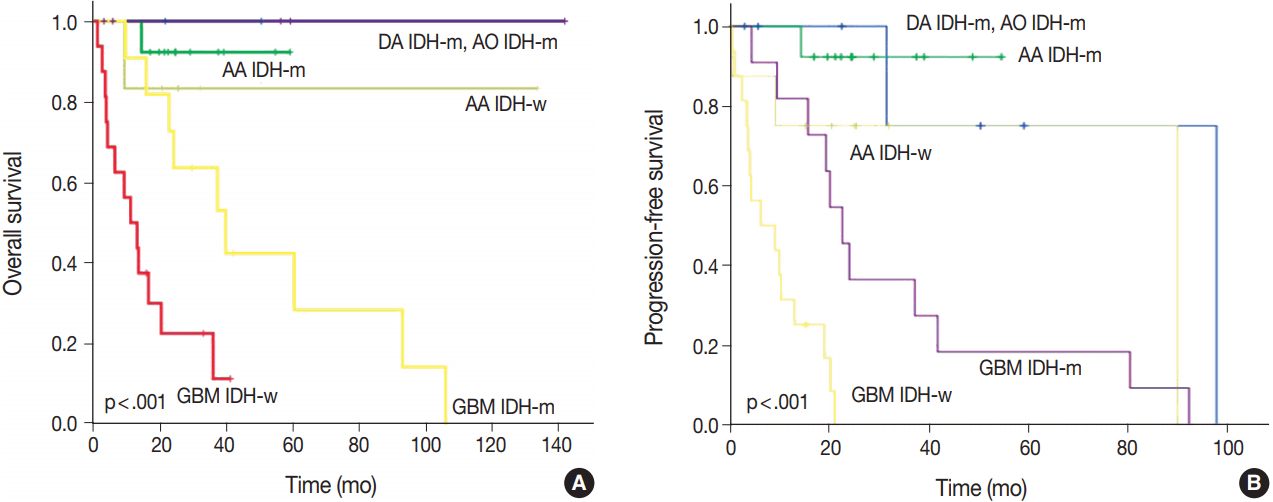
Table 1.
Demographic summary of the patient cohort according to the original diagnoses in this study
|
Parameter |
OA (n = 15) |
AOA (n = 14) |
GBMO (n = 29) |
|
Age (yr) |
|
|
|
|
Mean (range) |
40.8 (14–72) |
41.1 (13–64) |
53.5 (31–77) |
|
Median |
36 |
35.5 |
56 |
|
Sex (male:female) |
11:4 |
5:9 |
22:7 |
|
Median OS (mo) |
45.3 |
21.7 |
26.7 |
|
Median PFS (mo) |
39.8 |
20.9 |
19.0 |
|
Tumor location |
F8 T4 P1 PT1 HIPP1 |
F6 T5 P1 O1 TH1 |
F13 T10 P1 TH1 CC1 PO1 FTI1 FTP1 |
|
Treatment |
Surgery only |
Surgery only 13 |
CCRT 21 |
|
|
Others 1 |
Others 8 |
Table 2.
IDH1-F/IDH1-R and IDH2-F/IDH2-R primers
|
Primer |
Sequence (5’ → 3’) |
|
IDH 1-F |
ACCAAATGGCACCATACGA |
|
IDH 1-R |
GCAAAATCACATTATTGCCAAC |
|
IDH 2-F |
GCTGCAGTGGGACCACTATT |
|
IDH 2-R |
TGTGGCCTTGTACTGCAGAG |
Table 3.
Change in diagnosis after applying the genetically integrated diagnostic criteria
|
Original diagnosis (n = 58) |
Re-diagnosis (n = 58)
|
|
Criteria |
No. (%) |
|
OA, grade II (n = 14) |
DA IDH-m |
2 (14.3) |
|
DA IDH-w |
2 (14.3) |
|
AA IDH-m |
7 (50) |
|
AA IDH-w |
2 (14.3) |
|
AO IDH-m and 1p/19q-codeleted |
1 (7.1) |
|
AOA, grade III (n = 14) |
AA IDH-m |
6 (42.9) |
|
AA IDH-w |
5 (35.7) |
|
AO IDH-m and 1p/19q-codeleted |
3 (17.6) |
|
GBMO, grade IV (n = 29) |
GBM IDH-w |
16 (55.2) |
|
GBM IDH-m |
11 (37.9) |
|
AO IDH-m and 1p/19q-codeleted |
2 (6.9) |
Table 4.
Demographic summary of our cohort according to genetics integrated reclassification
|
Parameter |
DA IDH-m (n = 2) |
DA IDH-w (n = 2) |
AA IDH-m (n = 13) |
AA IDH-w (n = 8) |
AO (n = 6) |
GBM IDH-m (n = 11) |
GBM IDH-w (n = 16) |
|
Age, mean (range, yr) |
37.5 (31–44) |
49.5 (32–67) |
39.3 |
39.9 (14–64) |
52.6 (35–69) |
42.3 (34–58) |
58.4 (31–77) |
|
Sex (male:female) |
2:0 |
1:1 |
9:4 |
3:5 |
3:3 |
7:4 |
13:3 |
|
ATRX (–), n (%) |
2 (100) |
1 (50) |
12 (92.3) |
7 (87.5) |
0 |
7 (63.6) |
5 (31.3) |
|
1p/19q codel |
0 |
0 |
0 |
0 |
6 |
0 |
0 |
|
Primary GBM |
|
|
|
|
|
7 |
14 |
|
Secondary GBM |
|
|
|
|
|
4 |
1 |
|
OS (mo) |
36.5 |
19.5 |
29.8 |
34.5 |
45.5 |
43.6 |
14.5 |
|
PFS (mo) |
36.0 |
18.9 |
29.0 |
27.3 |
34.0 |
33.4 |
9.1 |
|
Tumor location |
F1 P1 |
F2 |
F5 T6 P1 PT1 |
F4 T1 O1 TH1 HIPP1 |
F4 T2 |
F6 T3 FTI1 FTP1 |
F5 T7 P1 CC1 PO1 TH1 |
|
Treatment |
Surgery only |
Surgery only |
Surgery only |
Surgery only 7 |
Surgery only 4 |
CCRT 10 |
CCRT 11 |
|
|
|
|
Others 1 |
CCRT 1 |
Others 1 |
Others 5 |
|
|
|
|
|
Others 1 |
|
|









 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download