This article has been
cited by other articles in ScienceCentral.
Abstract
Background:
As measurement of Ki-67 proliferation index is an important part of breast cancer diagnostics, we conducted a multicenter study to examine the degree of concordance in Ki-67 counting and to find factors that lead to its variability.
Methods:
Thirty observers from thirty different institutions reviewed Ki-67–stained slides of 20 different breast cancers on whole sections and tissue microarray (TMA) by online system. Ten of the 20 breast cancers had hot spots of Ki-67 expression. Each observer scored Ki-67 in two different ways: direct counting (average vs. hot spot method) and categorical estimation. Intraclass correlation coefficient (ICC) of Ki-67 index was calculated for comparative analysis.
Results:
For direct counting, ICC of TMA was slightly higher than that of whole sections using average method (0.895 vs 0.858). The ICC of tumors with hot spots was lower than that of tumors without (0.736 vs 0.874). In tumors with hot spots, observers took an additional counting from the hot spot; the ICC of whole sections using hot spot method was still lower than that of TMA (0.737 vs 0.895). In categorical estimation, Ki-67 index showed a wide distribution in some cases. Nevertheless, in tumors with hot spots, the range of distribution in Ki-67 categories was decreased with hot spot method and in TMA platform.
Conclusions:
Interobserver variability of Ki-67 index for direct counting and categorical estimation was relatively high. Tumors with hot spots showed greater interobserver variability as opposed to those without, and restricting the measurement area yielded lower interobserver variability.
Keywords: Ki-67, Observer variation, Multicenter, Breast neoplasms
Breast cancer is a heterogeneous group of diseases with different molecular bases, histologies, and prognoses [
1]. Current classification is based on molecular subtyping with cDNA microarrays into five categories: luminal A, luminal B, human epidermal growth factor receptor 2 (HER2)–positive, basal-like, and normal breast-like [
2,
3]. In daily practice, however, immunohistochemistry is a simple and economic method that provides as much valuable information as gene expression profiling. Thus, breast cancer subtyping using immunohistochemical surrogates has been proposed [
4-
6]. Cheang
et al. [
6] classified breast cancers into luminal A, luminal B, HER2-positive, and triple negative using estrogen receptor, progesterone receptor, HER2, and Ki-67. They used a 14% cutoff value for Ki-67 with hormone receptor–positive breast cancer having Ki-67<14% as luminal A and those with ≥14% as luminal B [
6]. The St. Gallen Consensus for treatment of early breast cancer adopted the surrogate definitions of breast cancer subtypes proposed by Cheang
et al. [
6] and established guidelines according to each subtype, which are now held standard of treatment in breast cancer [
7,
8]. Because classification of luminal A and luminal B subtypes depends on Ki-67 index, its accurate measurement is a critical factor in deciding therapeutic modalities.
Ki-67 is a protein expressed in all phases of the cell cycle (except for resting Go) used as a tumor proliferation marker [
9]. Ki-67 index is defined by the percentage of tumor cells with positive nuclear staining out of all tumor cells within a given histological field. Several studies have shown the prognostic and predictive value of Ki-67 with a high Ki-67 index having higher risks of recurrence and treatment response in breast cancer [
10-
13]. Furthermore, a more recent study has shown that for neoadjuvant endocrine therapy, Ki-67 indices before treatment and 2 weeks after treatment are strongly associated with time to relapse as well as response to endocrine treatment [
14]. Despite such critical role of Ki-67 index in breast cancer treatment, there exist dilemma in using it as a standard prognostic or predictive marker owing to the lack of standard methodologies in its measurement.
There are some methodological issues concerning Ki-67 index interpretation, which can cause variabilities in its measurement. First, the number of tumor cells counted and the areas selected for counting may be different for each observer. Second, as with other tumors, breast cancers may exhibit intratumoral heterogeneity in cell proliferation with areas of higher proliferation indicated by more intense staining of Ki-67 known as “hot spots.” In tumors with such hot spots, an observer may decide to measure Ki-67 index from the hot spot or take the average of a few different fields in the same section. Third, some may proceed with manual counting while others may utilize digital image analysis (DIA). Fourth, some may record Ki-67 index in continuous numbers while others may record it in categorical values. Together, these various factors inevitably lead to interobserver variability in Ki-67 measurement, the degree of which is unknown. Thus, suggestions have been put forth by the International Ki-67 in Breast Cancer Working Group in an attempt to standardize Ki-67 measurement [
15]. The need for standard criteria regarding the number of tumor cells and the area of the field to be counted, as well as the method of counting is becoming all the more urgent.
In this study, we chose 20 samples of invasive breast cancer, of which 10 had hot spots and performed a multicenter study with participation from 30 institutions to assess the degree of interobserver variability in Ki-67 index measurement and clarify the factors affecting its measurement by comparing whole sections versus tissue microarray (TMA) platforms, average counting versus hot spot counting, and continuous versus categorical counting.
MATERIALS AND METHODS
Case selection and TMA construction
Twenty cases (T1–T20) of invasive ductal carcinoma (IDC) were selected from the pathologic archives of Seoul National University Bundang Hospital after reviewing the Ki-67 immunostained slides upon diagnosis. All cases were surgically resected tissue specimens fixed in 10% formalin and paraffin-embedded in 2012. To analyze the effect of the presence of hot spots in Ki-67 measurement, we purposely chose 10 IDCs (T11–T20) which had localized high Ki-67 staining areas (
Fig. 1). The remaining (T1–T10) showed a relatively even distribution of positive tumor cells. All twenty cases were arranged into a 2 mm, single-core TMA to set limits for the area of analysis, as opposed to whole section slides. TMA was constructed using a trephine apparatus (Superbiochips Laboratories, Seoul, Korea). Hot spot areas were selected to be incorporated into the TMA in 10 IDCs (T11–T20) while areas most representative of the tumor were selected for the remaining 10 cases (T1–T10) for TMA construction. This study was exempt from Institutional Review Board (IRB) deliberation (IRB No. X-1508/312-901).
Immunohistochemical staining and measurement of Ki-67
Immunohistochemical staining of Ki-67 was re-performed using MIB-1 clone (1:500; DAKO, Carpinteria, CA, USA) in whole sections and a TMA section. Four-micrometer-thick tissue sections were cut, dried, deparaffinized, and rehydrated following standard procedures. All sections were subjected to heat-induced antigen retrieval. Immunohistochemical staining was carried out in a BenchMark XT autostainer (Ventana Medical Systems, Tucson, AZ, USA) using an UltraView detection kit. All immunostained slides were scanned using ScanScope CS system (Aperio, Vista, CA, USA) and the files of such scanned slides were uploaded to an online system. Thirty different pathologists (O1–O30) from thirty different institutions (25 of which are academic teaching hospitals including all of the major hospitals in the country) participated in this study and examined the scanned slides using ImageScope viewing software with specific guidelines for Ki-67 measurement.
All observers measured Ki-67 index in two different ways for both whole section slides and a TMA slide. First, direct counting method consisted of counting at least 500 tumor cells in each of three representative areas in whole sections and counting of at least 500 tumor cells or all of the tumor cells (if the number of tumor cells were less than 500) in a TMA, either manually or digitally using an image analyzer, and the positive percentage was calculated; two out of 30 participants utilized DIA for direct counting. Second, rough estimation method consisted of visual estimation of Ki-67 index into five categories: 0 (<5%), 1 (5% to <10%), 2 (10% to <20%), 3 (20% to <50%), and 4 (≥50%). In tumors with hot spot areas (T11–T20), direct counting on whole section slides consisted of two different methods. The first was to count the number of positive tumor cells in one representative hot spot area which we designated as the “hot spot method.” The other method was counting in three representative areas including the hot spot and then calculating its average, which we named as the “average method.” For rough estimation, the observers made separate categorical estimations of Ki-67 from the hot spot and the entire field in whole section slides in tumors with hot spot areas (T11–T20). The schematic representation of study design and counting methods is provided in
Fig. 2.
Statistical analyses
To assess interobserver variability in the 20 cases, we calculated intraclass correlation coefficient (ICC) with a 95% confidence interval (CI). Since there is no consensus on standard criteria for ICC, we used this value only for comparison of interobserver variabilities of different Ki-67 measurement methods. The ICC has a range of 0 to 1, with1 defining perfect agreement. To evaluate the distribution of Ki-67 proliferation indices measured by different observers for each case, we calculated the mean, median, standard error of mean, and standard deviation of each case, for every method. We drew a side by side box plot for Ki-67 indices. All data analyses were performed using IBM SPSS ver. 21.0.0 (IBM Co., Armonk, NY, USA).
RESULTS
Direct counting
The Ki-67 indices in each tumor calculated by direct counting in whole sections and a TMA are presented in
Fig. 3. The mean and median values of Ki-67 indices were relatively lower in T1–T10 than in T11–T20 for both whole sections and TMA. To evaluate the effect of restricting area of measurement on Ki-67 counting, we compared the ICCs of Ki-67 indices in whole sections and TMA. The ICC of 20 cases among 30 observers was 0.802 (95% CI, 0.696 to 0.897) for whole sections by the average method. The ICC of Ki-67 indices measured in TMA was 0.895 (95% CI, 0.829 to 0.948). Because the TMA was constructed from the hot spot areas in T11–T20, we also compared the Ki-67 indices acquired by hot spot method in whole sections with those in TMA (
Table 1). In T11–T20, the ICC of TMA was higher than that of whole sections using hot spot method (TMA: ICC, 0.895 [95% CI, 0.798 to 0.966]; whole section: ICC, 0.737 [95% CI, 0.561 to 0.905]).
To assess the effect of the presence of hot spots on direct counting, we compared the ICC of tumors without hot spots (T1–T10) with that of tumors with hot spots (T11–T20). Ki-67 indices using the average method were used for analysis in both T1–T10 and T11–T20. The ICC of T1–T10 was 0.874 with a 95% CI of 0.761–0.959, and the ICC of T11–T20 was 0.736 with a 95% CI of 0.559–0.904 in whole sections. The ICC of tumors with hot spots was significantly lower than that of tumors without hot spots using the same method of counting.
We also compared the ICC of two different counting methods applied in T11–T20: hot spot method versus average method (
Table 1). In general, the mean Ki-67 indices resulting from the hot spot method was higher than those yielded using the average method. However, there was no difference between the ICC value of the former (ICC, 0.737 [95% CI, 0.561 to 0.905] and latter ICC, 0.736 [95% CI, 0.559 to 0.904]).
Categorical estimation
The Ki-67 indices in each tumor calculated by rough estimation for whole sections and TMAs are presented in
Fig. 4. In whole sections, Ki-67 indices belonged to two categories in six cases, three categories in six cases, four categories in seven cases, and five categories in the remaining one case. In TMAs, they were assigned to one category in one case, two in six, three in eight, and four in the remaining five cases. Ki-67 for T15 in particular, showed a poor concordance between observers in the whole section. One observer assessed Ki-67 index as less than 5% while another assessed it as more than 50%. Rough estimation in TMA resulted in a narrower distribution of Ki-67 categories in six cases (T11 and T15 through 20) and a wider distribution of Ki-67 in three cases (T7, T10, and T14) compared with that in whole sections. Based on the distribution of Ki-67 indices, the rough estimation method was not better than the direct counting method.
Additionally, we compared the Ki-67 indices acquired by rough estimation in whole section using hot spot method with those in TMA for T11–T20 (
Table 2). The distribution of Ki-67 categories was mainly restricted to two continuous categories in both platforms. In T11–T20, we also compared the Ki-67 indices acquired by categorical estimation in two different ways: hot spot method versus average method (
Table 2). We observed that tumors tended to be categorized into higher categories and the range of distribution was decreased using the hot spot method compared with the average method.
DISCUSSION
Although Ki-67 index in breast cancer has predictive and prognostic values, it has not been accepted as a standard predictive or prognostic biomarker due to variability in its measurement. Thus, in this study, we performed a multicenter study to assess the degree of interobserver variability in Ki-67 index measurement and to find factors that contribute to its variability.
As there is no statistical criterion defining “high” concordance, one study assumed it to be having an ICC of approximately 0.9 [
16]. The ICCs for various measurements taken in our study were in the range of 0.7–0.8. Thus, interobserver concordance in Ki-67 measurement was moderately good at best for all settings. We were, however, able to deduce some important conclusions from our study. With the participation of thirty different institutions, we evaluated interobserver variability in Ki-67 index measurement specifically depending on the area of selection, intratumoral heterogeneity, and counting method, which we held especially important among several factors that are known to influence Ki-67 measurement variability.
Previous studies evaluating interobserver variability of Ki-67 measurement have used either core biopsy samples or TMAs. However, this study is the first one, to our best knowledge, to have made direct comparisons between the whole section and TMA preparations from the same tumor sample. While restricting the area of counting increased concordance between the observers by a small amount (TMA: ICC, 0.895 [95% CI, 0.829 to 0.948]; whole section: ICC, 0.858 [95% CI, 0.774 to 0.929]), the difference was not significantly large. However, in tumors with hot spots (T11–T20), the ICC of TMA was much higher than that of whole sections using either average method or hot spot method (TMA: ICC, 0.895 [95% CI, 0.798 to 0.966]; whole section, average method: ICC, 0.736 [95% CI, 0.559 to 0.904]; whole section, hot spot method: ICC, 0.737 [95% CI, 0.561 to 0.905]). Similarly, in categorical estimation, the range of distribution in Ki-67 categories was decreased when counting in TMA in the cases with hot spots. Thus, restricting the area of counting seems to be a critical factor affecting concordance in Ki-67 measurement.
Because some breast cancers possess intratumoral heterogeneity in tumor cell proliferation, a manifestation of which is the presence of hot spots in Ki-67 expression, we attempted to analyze the degree of difference in Ki-67 measurement between observers in two ways. Dividing those cancers with and without hot spots and measuring ICCs of the two groups for the average counting method yielded an ICC of 0.874 for T1–T10 (95% CI, 0.761 to 0.959) and 0.736 (95% CI, 0.559 to 0.904) for T11–T20 in whole sections. Thus, the ICC of tumors with hot spots was much lower than that of tumors without hot spots; such finding that tumors with hot spots exhibit greater interobserver variability is not counterintuitive. A more interesting finding is that within this subset of tumors with hotspots, when we measured the degree of difference in Ki-67 measurement with regards to counting an average of the entire field or taking a single count from the hot spot only, there was no difference between the ICC value of the former (ICC, 0.736 [95% CI, 0.559 to 0.904]) and latter (ICC, 0.737 [95% CI, 0.561 to 0.905]). We expected that the ICC for whole section using hot spot method would be higher than that for whole section using average method. However, even within the hot spots, it was likely that observers chose different areas for counting, and the distribution of Ki-67–stained tumor nuclei was variable.
Interpretation of the weakly stained nuclei as either positive or negative staining may be an important factor contributing to variability in Ki-67 measurement. However, we did not investigate this interpretative issue in the current study. We used a mouse monoclonal antibody MIB-1 for Ki-67 immunostaining. It yielded similar yet less intense staining results compared with rabbit monoclonal antibody SP6 using the central laboratory equipment (data not shown). SP6 was employed in many previous pivotal studies [
6,
17-
20] and showed increased sensitivity compared to mouse monoclonal antibodies [
17,
18,
20]. However, the current recommendations from the International Ki-67 in Breast Cancer Working Group states only MIB-1 as the antibody endorsed in Ki-67 measurement [
15,
21]. Further comparative studies using different clones for Ki-67 and interpretation of weakly stained nuclei will be needed.
In order to assess interobserver variability depending on the counting method (in categories or direct numbers), each observer was instructed to take these two counting methods for each sample: for direct counting, more than five hundred cells were counted from at least three representative areas. For categorical counting, the observers made an estimate of the percentage of positive tumor cells into five categories (category 0, <5%; category 1, 5%–10%; category 2, 10%–20%; category 3, 20%–50%; and category 4, ≥50%). Although Ki-67 indices belonged to two categories in six cases on whole section and TMA (one case on TMA belonged to only one category), they had a wide distribution belonging to more than three categories in the remaining cases. Moreover, the cases with Ki-67 index within 10%–20%, which includes the cutoff value of 14% used for distinction of luminal A and luminal B subtypes in breast cancer, showed wide distribution in rough estimation from category 0 to 4. Therefore, rough estimation methods may be worse than direct counting method in terms of interobserver concordance.
A limitation of our study includes a small number of tumor samples. Although each tumor sample was presented in two platforms of whole section and TMA, and each observer made at least a few countings for each sample for various settings, the absolute sample number was only twenty, which is the major reason we restricted our statistical analysis to evaluating ICCs.
Finally, all of the observers were given the liberty of using either manual or digital counting, and two observers made direct counting via DIA. However, we were not able to make direct comparisons between the two methods due to the small sample size. DIA enables a much quicker measurement, and its clinical utility seems promising. One group has reported that automated Ki-67 measurement was almost as comparable as visual estimation in precision and prognostic ability [
22], and Laurinavicius
et al. [
23] have suggested specific methodologies in using DIA for Ki-67 measurement. Standardization of DIA techniques may offer more consistent results between observers in the future, and its application using standard protocols should be studied in further studies.
In conclusion, our nationwide thirty-center study of Ki-67 interobserver variability showed that interobserver variability in measuring this critical biomarker is high. Although direct comparison between direct counting and rough estimation into categories was impossible due to statistical issues, direct counting seems to result in less interobserver variability. Thus, we recommend that Ki-67 measurement be performed by direct counting rather than by rough categorical estimation. Tumors with hot spots generally showed greater interobserver variability than those without hot spots. However, restricting the area of measurement to TMA platform resulted in decreased interobserver variability, even in tumors with hot spots. At this point, we cannot put down a specific number for the area of selection, and further studies are needed; however, we can propose that the area of selection should be confined to a specific area, such as the periphery of the tumor or hot spots (in cases with hot spots). Lastly, we urge future analyses comparing manual counting versus DIA in Ki-67 measurement since DIA seems to reduce not only intraobserver variability but also interobserver variability so that this critical biomarker can be used with a greater confidence in clinical practice.
ACKNOWLEDGMENTS
This study was supported by Korean Society of Pathologists Grant 2013, and by a grant from the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (Grant No. NRF-2015R1A2A2A01007907) to Park SY.
REFERENCES
1. Polyak K. Breast cancer: origins and evolution. J Clin Invest. 2007; 117:3155–63.

2. SØrlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001; 98:10869–74.
3. Perou CM, SØrlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000; 406:747–52.

4. Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004; 10:5367–74.

5. Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006; 295:2492–502.

6. Cheang MC, Chia SK, Voduc D, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009; 101:736–50.

7. Goldhirsch A, Winer EP, Coates AS, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013; 24:2206–23.
8. Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thurlimann B, Senn HJ. Strategies for subtypes: dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011; 22:1736–47.
9. Bruno S, Darzynkiewicz Z. Cell cycle dependent expression and stability of the nuclear protein detected by Ki-67 antibody in HL-60 cells. Cell Prolif. 1992; 25:31–40.

10. Luporsi E, André F, Spyratos F, et al. Ki-67: level of evidence and methodological considerations for its role in the clinical management of breast cancer: analytical and critical review. Breast Cancer Res Treat. 2012; 132:895–915.

11. Yerushalmi R, Woods R, Ravdin PM, Hayes MM, Gelmon KA. Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol. 2010; 11:174–83.

12. Viale G, Giobbie-Hurder A, Regan MM, et al. Prognostic and predictive value of centrally reviewed Ki-67 labeling index in post-menopausal women with endocrine-responsive breast cancer: results from Breast International Group Trial 1-98 comparing adjuvant tamoxifen with letrozole. J Clin Oncol. 2008; 26:5569–75.

13. Penault-Llorca F, André F, Sagan C, et al. Ki67 expression and docetaxel efficacy in patients with estrogen receptor-positive breast cancer. J Clin Oncol. 2009; 27:2809–15.

14. Dowsett M, Smith IE, Ebbs SR, et al. Prognostic value of Ki67 expression after short-term presurgical endocrine therapy for primary breast cancer. J Natl Cancer Inst. 2007; 99:167–70.

15. Dowsett M, Nielsen TO, A’Hern R, et al. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst. 2011; 103:1656–64.
16. Polley MY, Leung SC, McShane LM, et al. An international Ki67 reproducibility study. J Natl Cancer Inst. 2013; 105:1897–906.
17. Wong SC, Chan JK, Lo ES, et al. The contribution of bifunctional SkipDewax pretreatment solution, rabbit monoclonal antibodies, and polymer detection systems in immunohistochemistry. Arch Pathol Lab Med. 2007; 131:1047–55.

18. Zabaglo L, Salter J, Anderson H, et al. Comparative validation of the SP6 antibody to Ki67 in breast cancer. J Clin Pathol. 2010; 63:800–4.

19. Ekholm M, Beglerbegovic S, Grabau D, et al. Immunohistochemical assessment of Ki67 with antibodies SP6 and MIB1 in primary breast cancer: a comparison of prognostic value and reproducibility. Histopathology. 2014; 65:252–60.

20. Fasanella S, Leonardi E, Cantaloni C, et al. Proliferative activity in human breast cancer: Ki-67 automated evaluation and the influence of different Ki-67 equivalent antibodies. Diagn Pathol. 2011; 6 Suppl 1:S7.

21. Colozza M, Azambuja E, Cardoso F, Sotiriou C, Larsimont D, Piccart MJ. Proliferative markers as prognostic and predictive tools in early breast cancer: where are we now? Ann Oncol. 2005; 16:1723–39.

22. Mohammed ZM, McMillan DC, Elsberger B, et al. Comparison of visual and automated assessment of Ki-67 proliferative activity and their impact on outcome in primary operable invasive ductal breast cancer. Br J Cancer. 2012; 106:383–8.

23. Laurinavicius A, Plancoulaine B, Laurinaviciene A, et al. A methodology to ensure and improve accuracy of Ki67 labelling index estimation by automated digital image analysis in breast cancer tissue. Breast Cancer Res. 2014; 16:R35.

Fig. 1.
A representative case with hot spots in Ki-67 immunohistochemistry. (A) Scan power view of Ki-67 immunostained slides with a hot spot in right lower corner. (B) High power view of the hot spot.
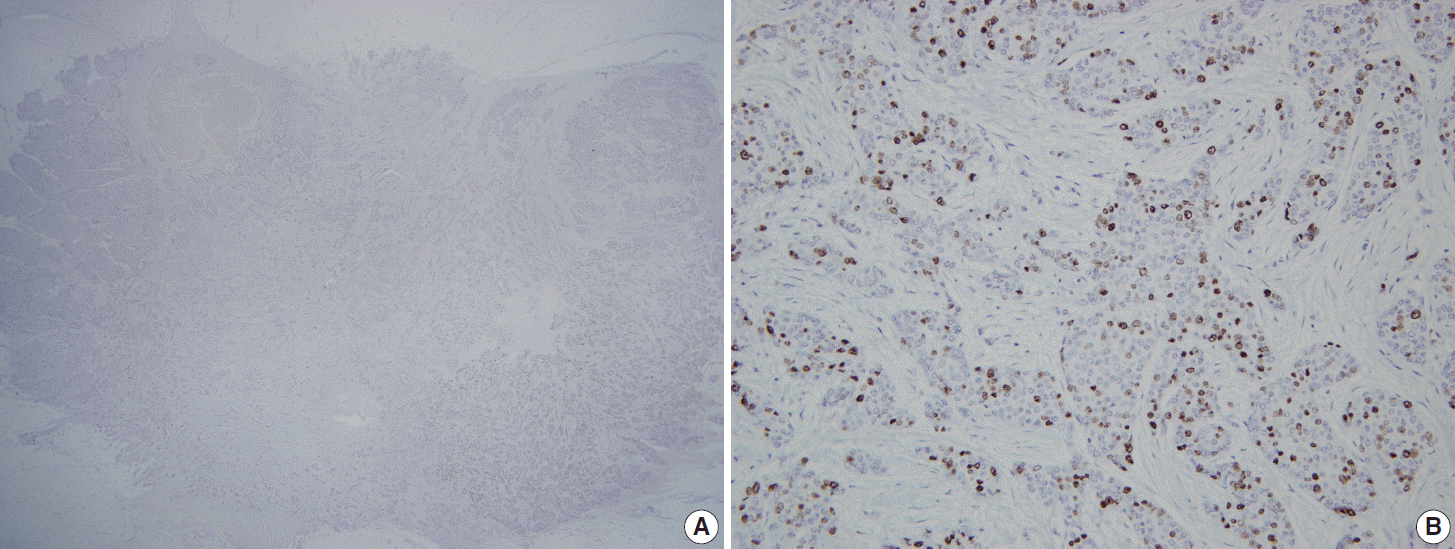
Fig. 2.
A schematic diagram of study design and counting methods. Twenty cases of invasive ductal carcinoma (IDC) were prepared in two platforms-whole sections and a tissue microarray (TMA), and then digitally scanned for analysis in online system. Each observer was instructed to measure Ki-67 index in two platforms in two different ways (direct counting or rough categorical estimation) employing the average method and hot spot method.
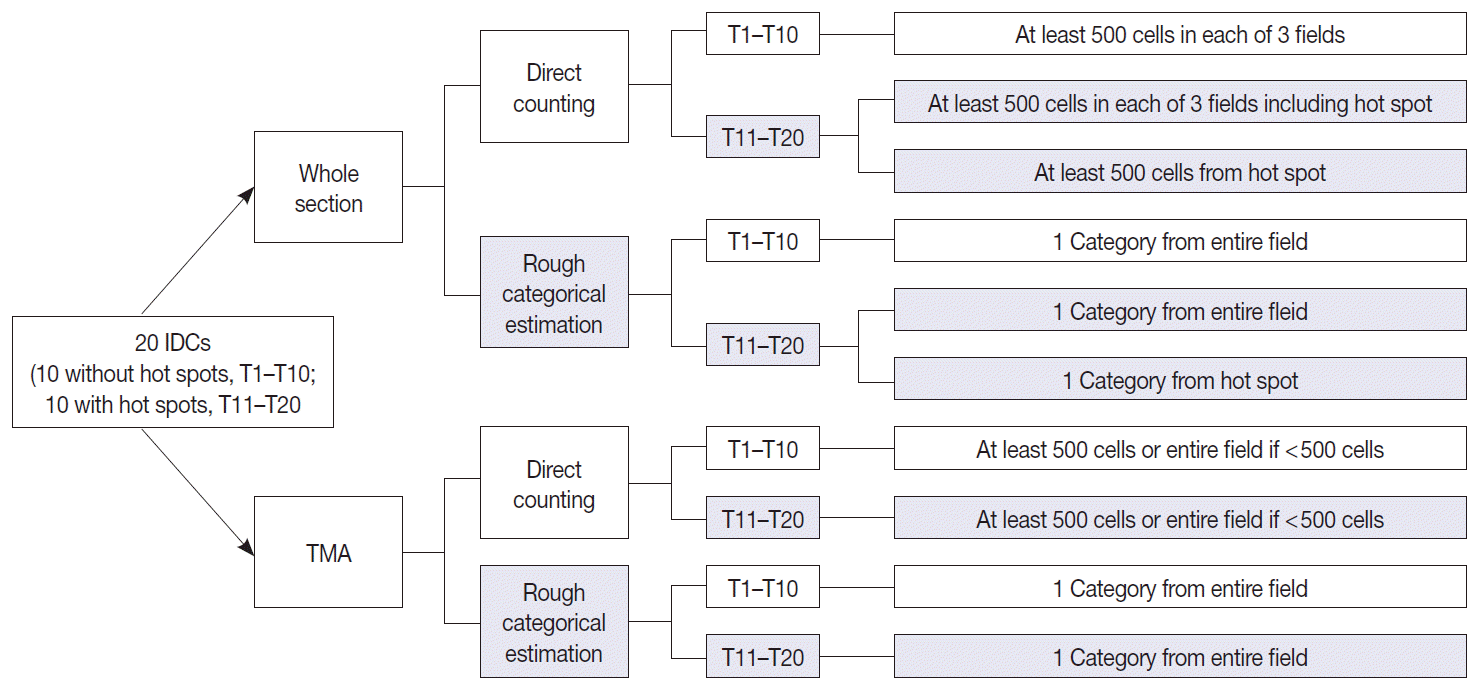
Fig. 3.
Side-by-side box plots of Ki-67 distribution using whole sections (A) and tissue microarray slides (B) in direct counting method. The box shows the first to third quartiles, the horizontal line inside the box represents the median, the whiskers extend to minimum and maximum values within 1.5 times the interquartile range (IQR) from the first and third quartiles. Outliers are represented by small circles and extreme values (more than 3 times IQR) by asterisks. The Ki-67 indices measured in whole section show wider distribution than those in tissue microarray.
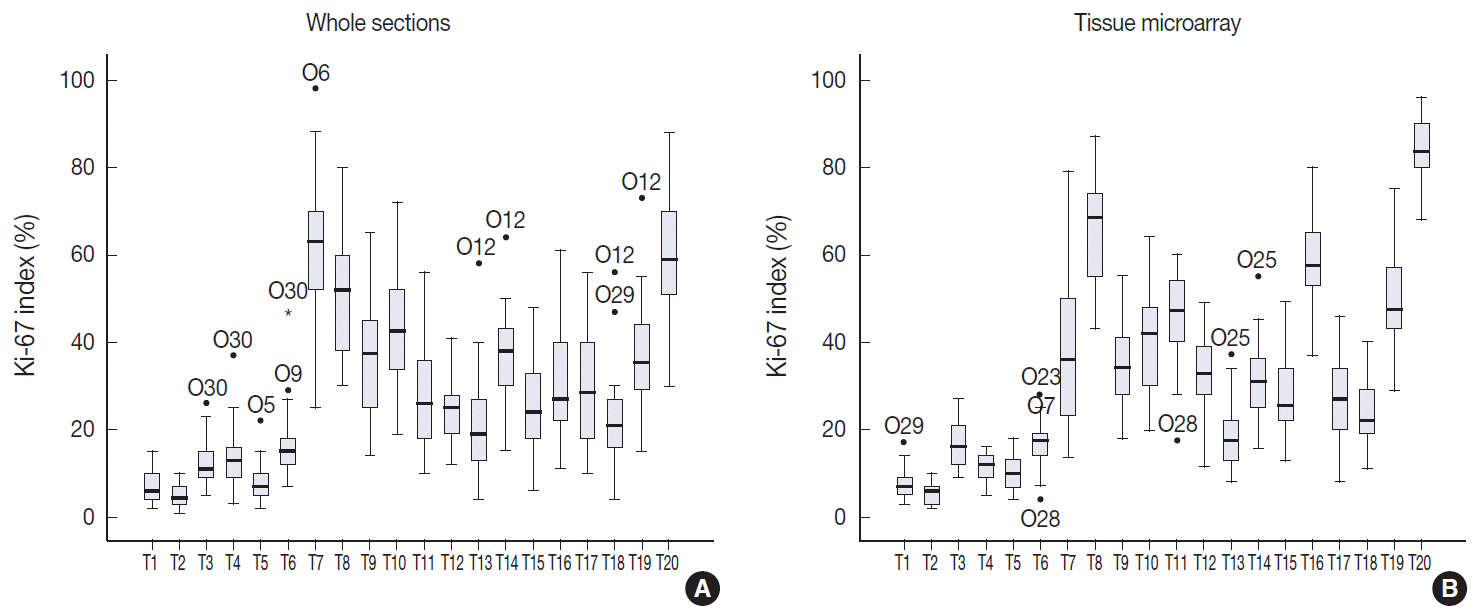
Fig. 4.
Distribution of Ki-67 indices in whole sections (A) and tissue microarray slides (B) in categorical estimation. The Ki-67 indices measured by categorical estimation in whole sections and TMAs show a wide distribution in some cases.
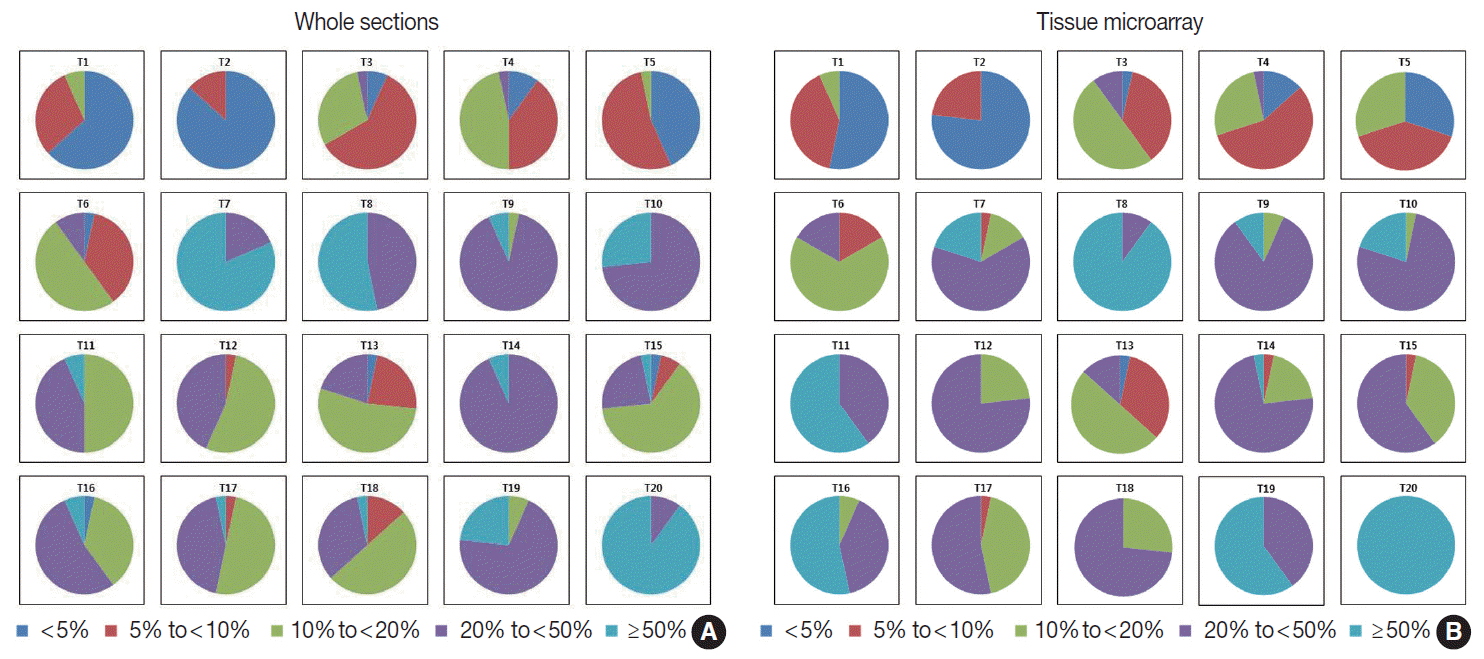
Table 1.
Comparison of Ki-67 assessment in whole sections and tissue microarray in T11–T20
|
T11 |
T12 |
T13 |
T14 |
T15 |
T16 |
T17 |
T18 |
T19 |
T20 |
|
Whole section, average method |
|
|
|
|
|
|
|
|
|
|
|
Minimum |
10.00 |
12.00 |
4.00 |
15.00 |
6.00 |
11.00 |
10.00 |
4.00 |
15.00 |
30.00 |
|
Maximum |
56.00 |
41.00 |
58.00 |
64.00 |
48.00 |
61.00 |
56.00 |
56.00 |
73.00 |
88.00 |
|
Mean |
28.20 |
24.43 |
20.50 |
37.20 |
24.97 |
31.30 |
29.27 |
21.93 |
37.13 |
59.90 |
|
Median |
26.00 |
25.00 |
19.00 |
38.00 |
24.00 |
27.00 |
28.50 |
21.00 |
35.50 |
59.00 |
|
SD |
12.50 |
7.12 |
11.77 |
10.09 |
10.24 |
13.08 |
12.76 |
10.58 |
12.74 |
14.68 |
|
SE |
2.28 |
1.30 |
2.15 |
1.84 |
1.87 |
2.39 |
2.33 |
1.93 |
2.33 |
2.68 |
|
Whole section, hot spot method |
|
|
|
|
|
|
|
|
|
|
|
Minimum |
24.80 |
23.50 |
13.50 |
20.60 |
17.50 |
27.00 |
15.80 |
16.50 |
33.30 |
62.50 |
|
Maximum |
62.20 |
56.00 |
36.50 |
59.50 |
61.00 |
79.30 |
54.50 |
49.00 |
72.50 |
94.00 |
|
Mean |
44.59 |
35.74 |
26.73 |
41.71 |
34.40 |
56.43 |
37.03 |
29.38 |
51.58 |
82.50 |
|
Median |
46.00 |
34.00 |
26.85 |
41.75 |
35.80 |
55.85 |
39.00 |
28.45 |
49.75 |
83.35 |
|
SD |
9.65 |
8.71 |
6.14 |
8.42 |
9.26 |
11.46 |
9.60 |
7.81 |
9.35 |
7.86 |
|
SE |
1.76 |
1.59 |
1.12 |
1.54 |
1.69 |
2.09 |
1.75 |
1.43 |
1.71 |
1.44 |
|
Tissue microarray |
|
|
|
|
|
|
|
|
|
|
|
Minimum |
17.50 |
11.60 |
8.00 |
15.80 |
13.00 |
37.00 |
8.00 |
11.00 |
29.00 |
68.00 |
|
Maximum |
60.00 |
49.00 |
37.00 |
55.00 |
49.00 |
80.00 |
46.00 |
40.00 |
75.00 |
96.00 |
|
Mean |
45.62 |
32.99 |
18.19 |
31.46 |
28.42 |
59.07 |
26.83 |
24.09 |
50.33 |
83.97 |
|
Median |
47.00 |
33.00 |
17.50 |
31.00 |
25.50 |
57.50 |
27.00 |
22.00 |
47.50 |
83.50 |
|
SD |
10.78 |
9.11 |
6.98 |
8.38 |
8.97 |
10.11 |
9.61 |
7.14 |
10.72 |
7.40 |
|
SE |
1.97 |
1.66 |
1.27 |
1.53 |
1.64 |
1.85 |
1.76 |
1.30 |
1.96 |
1.35 |
Table 2.
Ki-67 index assessment by rough estimation method in whole sections and tissue microarray in T11–T20
|
T11 |
T12 |
T13 |
T14 |
T15 |
T16 |
T17 |
T18 |
T19 |
T20 |
|
Whole section, average method |
|
|
|
|
|
|
|
|
|
|
|
< 5% |
0 |
0 |
1 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
|
5 to < 10% |
0 |
1 |
7 |
0 |
2 |
0 |
1 |
4 |
0 |
0 |
|
10 to < 20% |
15 |
16 |
16 |
0 |
19 |
11 |
15 |
15 |
2 |
0 |
|
20 to < 50% |
13 |
13 |
6 |
28 |
7 |
16 |
13 |
10 |
21 |
3 |
|
≥ 50% |
2 |
0 |
0 |
2 |
1 |
2 |
1 |
1 |
7 |
27 |
|
Whole section, hot spot method |
|
|
|
|
|
|
|
|
|
|
|
< 5% |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
5 to < 10% |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
10 to < 20% |
0 |
4 |
8 |
0 |
6 |
1 |
0 |
6 |
0 |
0 |
|
20 to < 50% |
22 |
24 |
20 |
15 |
22 |
8 |
20 |
22 |
8 |
0 |
|
≥ 50% |
8 |
2 |
1 |
15 |
2 |
21 |
10 |
2 |
22 |
30 |
|
Tissue microarray |
|
|
|
|
|
|
|
|
|
|
|
< 5% |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
5 to < 10% |
0 |
0 |
10 |
1 |
1 |
0 |
1 |
0 |
0 |
0 |
|
10 to < 20% |
0 |
7 |
15 |
6 |
11 |
2 |
13 |
8 |
0 |
0 |
|
20 to < 50% |
12 |
23 |
4 |
22 |
18 |
12 |
16 |
22 |
12 |
0 |
|
≥ 50% |
18 |
0 |
0 |
1 |
0 |
16 |
0 |
0 |
18 |
30 |








 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download