Abstract
Recent advances in optical molecular imaging allow identification of morphologic and biochemical changes in tissues associated with gastrointestinal (GI) premalignant lesions earlier and in real-time. This focused review series introduces high-resolution imaging modalities that are being evaluated preclinically and clinically for the detection of early GI cancers, especially Barrett esophagus and esophageal adenocarcinoma. Although narrow band imaging, autofluorescence imaging, and chromoendoscopy are currently applied for this purpose in the clinic, further adoptions of probe-based confocal laser endomicroscopy, high-resolution microendoscopy, optical coherence tomography, and metabolomic imaging, as well as imaging mass spectrometry, will lead to detection at the earliest and will guide predictions of the clinical course in the near future in a manner that is beyond current advancements in optical imaging. In this review article, the readers will be introduced to sufficient information regarding this matter with which to enjoy this new era of high technology and to confront science in the field of molecular medical imaging.
The best strategy to improve gastrointestinal (GI) cancer survival might be the use of feasible screening techniques and treatment of early lesions long before endoscopic diagnosis and the onset of overt clinical symptoms. As exemplified in esophageal adenocarcinoma, the early detection of Barrett esophagus (BE) with low- or high-grade dysplasia (HGD) might represent the most attractive goal to achieve a cure. This goal is aided by simultaneous advances in endoscopic treatment modalities such as submucosal endoscopic dissection and endoscopic ablation. This is especially true because currently, endoscopy alone is not able to clearly discriminate BE from columnar-lined epithelium or from areas of low-grade dysplasia, despite the development of chromoendoscopy, narrow band imaging (NBI), and high yield white-light endoscopy.1 Since BE is a precursor to esophageal adenocarcinoma and because late-stage adenocarcinoma is associated with a poor prognosis, endoscopic surveillance is recommended for patients with BE to enable early detection of neoplastic changes. For this surveillance, current guidelines recommend that random 4-quadrant biopsies be performed every 1 to 2 cm throughout the Barrett segment. However, this technique samples only a small portion of the epithelium and has been shown to miss areas of endoscopically ambiguous unapparent BE.2
Recent efforts have focused on developing novel diagnostic imaging technologies to detect the subtle epithelial changes associated with dysplasia and neoplasia in BE.3 Based on a meta-analysis, advanced imaging techniques such as chromoendoscopy or virtual chromoendoscopy significantly increase the diagnostic yield for dysplasia or cancer in patients with BE.4 As an example of the first advancement in molecular imaging technology, Sturm et al.5 have developed a peptide that binds specifically to BE presenting with HGD and BE-associated adenocarcinoma. They first applied the peptide ex vivo to esophageal specimens from 17 patients to validate specific binding and performed confocal endomicroscopy in vivo in 25 human subjects after topical peptide administration. They found 3.8-fold greater fluorescence intensity for esophageal neoplasia compared to BE and squamous epithelium with 75% sensitivity and 97% specificity. Since no toxicities were attributed to the peptide in either animal or patient studies, these first-in-human results concluded that this targeted imaging agent was quite safe and may be useful for guiding tissue biopsies as well as the early detection of BE. A second achievement in molecular imaging came from the concept of stem cell biology. The strategy by von Holzen and Enders6 and Franks7 was to generate stem cell clones from BE biopsy specimens in order to compare their gene expression profiles with patient-matched stem cell clones from esophageal squamous epithelia. By mining expression data from these Barrett's stem cell clones, they identified cell surface markers unique to the Barrett's stem cells, which could be recognized by cytotoxic antibodies. These findings can lead to the development of aptamers and can be used to help the endoscopist identify regions of atypia for biopsy, perform real-time diagnosis, stratify patients during the examination, and, ultimately, direct therapy in a pre-emptive manner to achieve the higher goal of complete removal of risky clones from BE, a strategy that will prevent future disaster.
The third approach for early molecular detection of BE involves recent important developments in biophotonics that have improved visualization of these subtle lesions sufficiently for cellular details to be seen in vivo during endoscopy. This allows for immediate and accurate diagnosis during the GI endoscopy, thereby avoiding the cost, risk, and time delay involved in biopsy and the several subsequent steps required for pathology analysis.8 Although chromoendoscopy, the Olympus NBI system (Olympus, Tokyo, Japan), high-yield white-light endoscopy, Fujinon (Fujinon Corp, Saitama, Japan) intelligent color enhancement, and point enhancement such as Pentax (Pentax, Tokyo, Japan) confocal laser endomicroscopy are examples of enhanced imaging technologies that are being used in daily practice, the combination of biophotonics with mass spectrometer imaging technology might enable detection and surveillance of upper GI early luminal malignancies, providing the imaging far beyond what is seeing. Combined with superimposing techniques such as chromoendoscopy or NBI and computed virtual chromoendoscopy with high-resolution/high-definition endoscopy, these advances in molecular imaging will highlight endoscopy as a tool for diagnosis as well as prevention. In this review article, further advances in these efforts to detect early GI cancer long before overt measurable clinical disease presents will be introduced and discussed, including advances in biomarker discovery, targeted imaging with novel fluorescent dye, next generation molecular imaging with proteomics, and imaging mass spectrometry (IMS) as exemplified in BE, a core premalignant lesion associated with esophageal adenocarcinoma.
Early diagnosis and minimally invasive treatment is the ideal target and every patient's desire, and even more importantly, these aspects are a matter of life or death for cancer patients. If tumors are detected as early as possible, complete treatment as well as minimally defective outcomes can be possible. Molecular diagnostics and the ability to screen tissue or blood samples for tumor specific genomic, proteomic, and epigenetic signatures might be required to achieve these improved outcomes. Therefore, the search for biomarkers that can detect the presence of tumors early in their development is essential. Once biomarkers in blood, urine, and other body fluids are discovered, molecular imaging can be used to detect tumors early and guide the selection of the optimal treatment modality, improving disease outcomes and survival. A number of blood-based biomarkers are already in clinical use for prognosis, response to treatment, and assessment of cancer progression, but in situ biomarkers can afford higher accuracy and additional information. It is interesting to question why scientists have not identified biomarkers capable of detecting cancer at an early stage with adequate sensitivity and specificity and where the bottleneck in this process is occurring. These questions can be answered by recent advances in proteomics approaches. Rapidly changing technologies for imaging, biomarker testing, and less-invasive endoscopic treatments benefit the patients and also promise to lower health system costs and reduce adverse events in patients.9 In a recent biomarker discovery study, Iwaya et al.10 showed reduced expression of α-N-acetylglucosamine in BE adjacent to BE-associated adenocarcinoma, α-methylacyl-CoA racemase as a predictor for neoplastic progression,11 p53 status as a biomarker for progression to cancer,12 and receptor tyrosine kinase EPHB4 as a carcinogenesis biomarker,13 among other results. As part of our efforts to identify biomarkers for BE, we have generated a BE rat model with esophagojejunostomy bypass surgery (Fig. 1A, arrow denotes the change in BE) and have performed cDNA microarray analysis to identify biomarkers for BE using tissues from the animal model (Fig. 1B). Using the critical genes identified by this high-throughput analysis, we are validating genes that may allow early diagnosis of BE long before overt appearance, as seen in BE patients by endoscopic diagnosis. Biomarker discovery could be used to optimize endoscopic surveillance strategies following ablation and to improve early detection.
In the near future, we may be able to detect HGD and resultant adenocarcinoma in patients with BE without an invasive diagnostic examination. Sturm et al.5 were very successful in their first-in-human testing of ASY*-FITC, conducted in accordance with good manufacturing practices. They showed promising results that their peptide was very safe and well tolerated in both humans and animals and produced bright illumination with easily detectable areas of esophageal adenocarcinoma and HGD, whereas BE was detectable to a minimal extent. The tissues identified as cancerous were confirmed via histology, showing that the labeled synthetic peptide called ASY*-FITC recognizes cancer tissue and enables in vivo imaging with a naked eye. Li et al.14 developed an affinity peptide, SNFYMPL, to detect dysplasia and high-risk BE in a larger number of patients with normal, BE, dysplasia, and cancer as well as normal gastric mucosa. These achievements highlight the current advances in confocal laser microscopy.15
Although it is possible to obtain hundreds of proteomic biomarker candidates, a high-throughput method for the development and refinement of selected reaction monitoring (SRM) assays is required for clinical application. For instance, a method must be applied to generate such assays for more than 1,000 cancer-associated proteins that, for instance, are functionally related to candidate cancer driver mutations. Through public access to the entire assay library, researchers will be able to use the detectability information in plasma and urine as a guide to target their cancer-associated proteins of interest in any sample type. This is why the generated expandable reference map of SRM assays for cancer-associated proteins will be a valuable resource for accelerating and planning biomarker verification studies, enabling cancer-associated biomarker validation.16 Multiple reaction monitoring (MRM) mass spectrometry (MS) is a similar tool that allows sensitive, precise quantitative analysis of peptides and the proteins from which they are derived. Key advantages of MRM assays are the ability to target specific peptide sequences, including variants and modified forms, and the capacity for multiplexing that allows analysis of dozens to hundreds of peptides.17 However, despite the advantages and potential, these techniques are limited because developing and validating MRM-MS-based assays is an extensive and iterative process that requires a coordinated and collaborative effort by the scientific community through the sharing of publicly accessible data and datasets, bioinformatics, and standard operating procedures by which the SRM has been created, which is a targeted MS technique emerging in the field of proteomics as a complement to untargeted shotgun methods.18 SRM is particularly useful when predetermined sets of proteins such as those constituting cellular networks or sets of candidate biomarkers need to be measured across multiple samples in a consistent, reproducible, and quantitatively precise manner. Seeing is not all that means throws the potential of these targeted or reaction monitoring-associated with proteomics to detect early premalignant lesions such as BE.
Rapid evaporative ionization mass spectrometry (REIMS) is an emerging technique that allows near real-time characterization of human tissue in vivo by analysis of the aerosol smoke released during electrosurgical dissection.19 The coupling of REIMS technology with electrosurgery for tissue diagnostics is known as the intelligent knife. Van den Broek et al.20 applied liquid chromatography/MS/MS for absolute quantification of ITIH4-derived putative biomarker peptides in clinical serum samples. They showed its applicability by quantifying all peptides in appropriate concentration ranges in serum from healthy volunteers as well as clinical samples from breast cancer patients. Recent advances in technology added a multifunctional semiclosed droplet-array chip coupled with electrospray ionization (ESI)-MS detection for multiple-sample pretreatment and analysis.21 A novel interfacing method for coupling the droplet system with ESI-MS was proposed using a sampling probe two-dimensional (2D) droplet array strategy. The additional uses of the semiclosed 2D droplet array and off-line interfacing mode might provide the system with the substantial flexibility and controllability in droplet indexing, multistep manipulating, and on demand sampling for MS analysis. No trials have been undertaken yet for BE, but we are now trying to apply this technology to BE tissues.
Although not a novel development, scanning spectral imaging techniques using scattered light have been applied as minimally invasive techniques to detect early cancerous changes in tissue and cell biology.22 Optical spectroscopic techniques have shown promising results in the diagnosis of disease on a cellular scale, leading to solid rationale for their use in the early detection of premalignant lesions. Since these techniques do not require any tissue removal or destruction, they can be applied to real-time diagnosis. Light scattering spectroscopy (LSS) relates the spectroscopic properties of light elastically scattered by small particles, such as epithelial cell nuclei and organelles, and is capable of characterizing the structural properties of tissue on a cellular and subcellular scale. However, to be useful for the detection of early cancerous changes that are otherwise not visible to the naked eye, it must rapidly survey a comparatively large area while simultaneously detecting these cellular changes. Perelman et al.23 demonstrated for the first time the successful LSS-based detection of dysplasia in BE using a simple proof of principle single-point instrument that is compatible with existing endoscopes, but with software and algorithms that provide additional quantitative and objective data about tissue structure and composition. In addition to endoscopic examination, a polarized scanning fiber optic probe is inserted into the endoscope working channel. Although this is not a new device, because entirely noninvasive devices capable of probing cellular composition on a subcellular scale for both structure and function are rather scarce, LSS revisited tools will provide unique capabilities to study functions of viable cells that are beyond the capabilities of other techniques and that are applicable to BE.24
Next generation sequencing allows the analysis of genomes, including those representing disease states, since genome-wide association studies revealed genomic risk loci that potentially have an impact on disease and phenotypic traits. However, the causes of most disorders are multifactorial, and systems level approaches, including the analysis of proteomes, are required for a more comprehensive understanding.25 The proteome is extremely multifaceted because of splicing and protein modifications, and this complex nature is further amplified by the interconnectivity of proteins into complexes and signaling networks. This complexity compels us to say good-bye to Western blotting.26 Because proteome analysis heavily relies on MS, MS based proteomics is starting to mature and to deliver through a combination of developments in instrumentation, sample preparation, and computational analysis. These developments are part of what is known as next generation proteomics, which, with additional application of molecular imaging, is being developed for the early detection of premalignant lesions.27
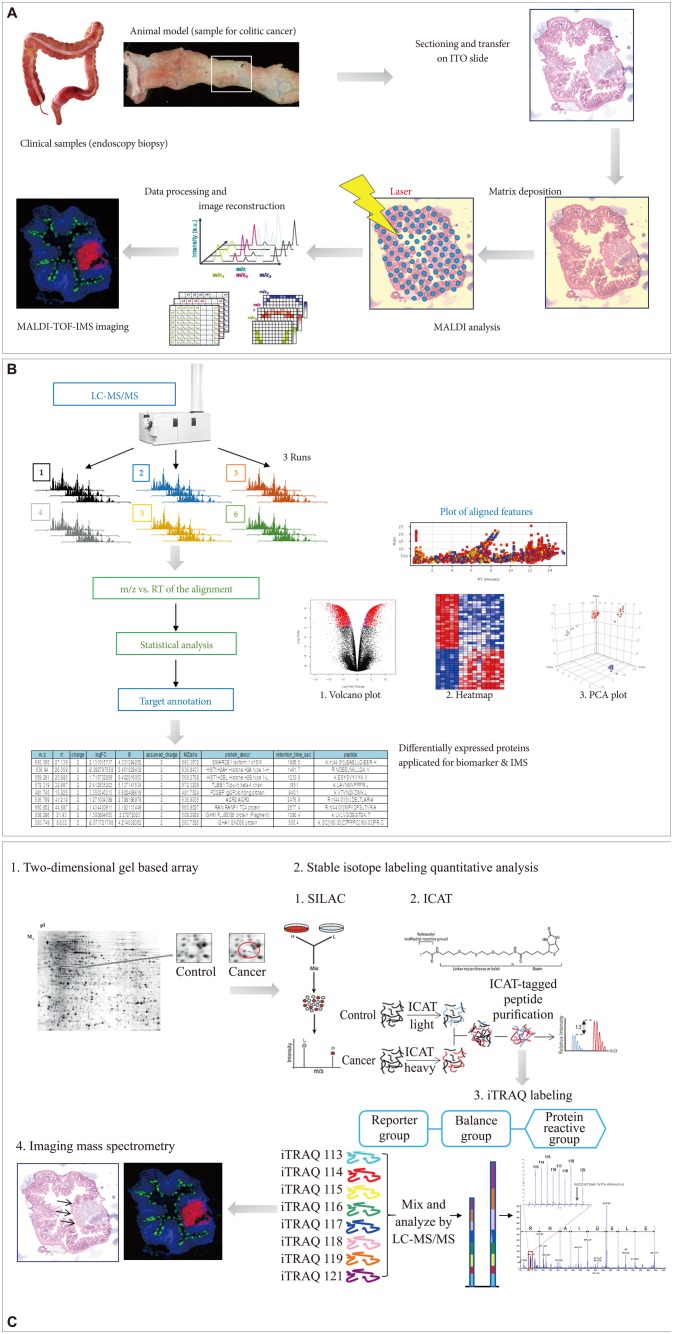
Proteomics has provided researchers with a sophisticated toolbox of labeling-based and label-free quantitative methods. Filiou et al.28 compared labeling-based and label-free quantitative proteomic techniques for clinical applications, assessed the use of labeled standards as internal controls for comparative studies in humans, and reviewed applications of labeling-based and label-free MS approaches in relevant model organisms and human subjects.29 They concluded that next generation proteomics might provide very useful insights into clinical disease pathogenesis and carcinogenesis, translating proteomics from the bench to the bedside. For instance, we have tried to identify potential biomarkers for disease discrimination or to predict the clinical course of inflammatory bowel disease, or to predict favorable response after Korean red ginseng intake using label-based isobaric tags for relative and absolute quantification (iTRAQ).30 Overall, it is clear that proteomics technologies are continuously evolving, and whether or not labeling is used is simply a matter of choice, because either way has advantages. The tremendous increase in proteome coverage over the past decade cannot be attributed to a single breakthrough, and there is ample scope for further developments. As seen in Fig. 2, we have tried to figure IMS imaging using the sample from chronic ulcerative colitis-associated colitic cancer. As shown in Fig. 2B, C, either label-free protein quantification (Fig. 2B) or label-based protein quantification (Fig. 2C) can be applied for both biomarker discovery and IMS imaging.
Current proteomic studies no longer focus only on identification of as many proteins as possible in a given sample, but instead try to accurately quantify them. Especially in clinical proteomics, the investigation of variable protein expression profiles can yield useful information on pathological pathways or drug targets relevant to certain clinical diseases. Popular quantitative strategies use either label-free approaches or stable isotope labeling. Label-free quantification through spectral counting and/or signal intensity of the detected peptides seems to be the least cumbersome way to obtain quantitative information, providing robust and precise relative protein expression information (Fig. 2B). However, care has to be taken with absolute copy number determinations of proteins.
Apart from label-free protein quantification, errors in quantification that occur due to variation in sample handling can be minimized when differential stable isotopes are introduced in the samples to be compared in order to create peptide isotopomers that can be distinguished in the output spectra based on their distinctive masses.31 Therefore, several strategies have been developed in recent years that use the incorporation of stable isotopes for which minimal variability is achieved using metabolic labeling in cell culture or even in whole organisms (Fig. 2C). Quantification is usually achieved at the MS level, except for chemical labeling using isobaric chemical labels, in which quantification is based on MS/MS reporter ions and quantification at the MS/MS level can be multiplexed, permitting the analysis of multiple perturbations in parallel.
Next-generation proteomics surely allows a much more in-depth view of the proteome in all its facets, and IMS technology provides benefits of see what was not seen and know what was not known, which is part of the wonderful world of modern medicine. As the core technology, MS will remain a main player in this arena. Combined with modern medicine tools, MS has evolved to be applicable in new drug development as well as digital pathology beyond anatomical pathology. Future advances in MSbased proteomics technologies will focus on 1) getting the relevant proteomics data with reduced analysis time, 2) reducing the quantity of material required, and 3) allowing indepth analysis of homogenous cell populations or micro-dissected tissue, with the ultimate aim being single-cell proteome analysis at the bedside or by human doctors. MS-based quantitative proteomics will become very powerful to discover disease biomarkers that are predictive or diagnostic, to identify prognostic and therapeutic targets, or to highlight translational medical research. All MS-based quantification strategies and techniques, including 2D gel-based methods, stable isotope labeling with amino acids in cell culture, isotope-coded affinity tag, and iTRAQ will contribute enormously to the early detection and prediction of disease long before it can be detected by white-light endoscopy (Fig. 2). With the advent of these advanced imaging technologies,32 BE can be a curable disease rather than being a precursor to esophageal cancer in the near future. To enjoy these advancements, the clinician should be equipped with sufficient knowledge about next generation proteomics, genomics, and sequencing. Routine integration will require the maturation and alignment of diverse postgenome technologies as well as crosstalk between different scientific communities.
References
1. Subramanian V, Ragunath K. Advanced endoscopic imaging: a review of commercially available technologies. Clin Gastroenterol Hepatol. 2013; 6. 28. Epub. DOI:10.1016/j.cgh.2013.06.015.

2. Chandra S, Gorospe EC, Leggett CL, Wang KK. Barrett's esophagus in 2012: updates in pathogenesis, treatment, and surveillance. Curr Gastroenterol Rep. 2013; 15:322. PMID: 23605564.

3. di Pietro M, Fitzgerald RC. Screening and risk stratification for Barrett's esophagus: how to limit the clinical impact of the increasing incidence of esophageal adenocarcinoma. Gastroenterol Clin North Am. 2013; 42:155–173. PMID: 23452636.
4. Qumseya BJ, Wang H, Badie N, et al. Advanced imaging technologies increase detection of dysplasia and neoplasia in patients with Barrett's esophagus: a meta-analysis and systematic review. Clin Gastroenterol Hepatol. 2013; 11:1562–1570. PMID: 23851020.

5. Sturm MB, Joshi BP, Lu S, et al. Targeted imaging of esophageal neoplasia with a fluorescently labeled peptide: first-in-human results. Sci Transl Med. 2013; 5:184ra61.

6. von Holzen U, Enders GH. A surprise cell of origin for Barrett's esophagus. Cancer Biol Ther. 2012; 13:588–591. PMID: 22549156.

7. Franks I. Barrett esophagus: new insights into the stem cell organization of Barrett esophagus. Nat Rev Gastroenterol Hepatol. 2012; 9:125. PMID: 22290494.
8. Templeton A, Hwang JH. Confocal microscopy in the esophagus and stomach. Clin Endosc. 2013; 46:445–449. PMID: 24143300.

9. Gordon LG, Mayne GC. Cost-effectiveness of Barrett's oesophagus screening and surveillance. Best Pract Res Clin Gastroenterol. 2013; 27:893–903. PMID: 24182609.

10. Iwaya Y, Hasebe O, Koide N, et al. Reduced expression of alphaGlcNAc in Barrett's oesophagus adjacent to Barrett's adenocarcinoma - a possible biomarker to predict the malignant potential of Barrett's oesophagus. Histopathology. Epub 2013 Oct 1. DOI: 10.1111/his.12296.
11. Kastelein F, Biermann K, Steyerberg EW, et al. Value of alpha-methylacyl-CoA racemase immunochemistry for predicting neoplastic progression in Barrett's oesophagus. Histopathology. 2013; 63:630–639. PMID: 24004067.
12. Fels Elliott DR, Fitzgerald RC. Molecular markers for Barrett's esophagus and its progression to cancer. Curr Opin Gastroenterol. 2013; 29:437–445. PMID: 23689523.

13. Hasina R, Mollberg N, Kawada I, et al. Critical role for the receptor tyrosine kinase EPHB4 in esophageal cancers. Cancer Res. 2013; 73:184–194. PMID: 23100466.

14. Li M, Anastassiades CP, Joshi B, et al. Affinity peptide for targeted detection of dysplasia in Barrett's esophagus. Gastroenterology. 2010; 139:1472–1480. PMID: 20637198.

15. Kiesslich R, Gossner L, Goetz M, et al. In vivo histology of Barrett's esophagus and associated neoplasia by confocal laser endomicroscopy. Clin Gastroenterol Hepatol. 2006; 4:979–987. PMID: 16843068.

16. Liebler DC, Zimmerman LJ. Targeted quantitation of proteins by mass spectrometry. Biochemistry. 2013; 52:3797–3806. PMID: 23517332.

17. Boja ES, Rodriguez H. Mass spectrometry-based targeted quantitative proteomics: achieving sensitive and reproducible detection of proteins. Proteomics. 2012; 12:1093–1110. PMID: 22577011.

18. Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods. 2012; 9:555–566. PMID: 22669653.

19. Liu YW, Aciego SM, Wanamaker AD Jr, Sell BK. A high-throughput system for boron microsublimation and isotope analysis by total evaporation thermal ionization mass spectrometry. Rapid Commun Mass Spectrom. 2013; 27:1705–1714. PMID: 23821564.

20. van den Broek I, Sparidans RW, Schellens JH, Beijnen JH. Sensitive liquid chromatography/tandem mass spectrometry assay for absolute quantification of ITIH4-derived putative biomarker peptides in clinical serum samples. Rapid Commun Mass Spectrom. 2010; 24:1842–1850. PMID: 20533314.

21. Su Y, Zhu Y, Fang Q. A multifunctional microfluidic droplet-array chip for analysis by electrospray ionization mass spectrometry. Lab Chip. 2013; 13:1876–1882. PMID: 23525283.

22. Qiu L, Turzhitsky V, Chuttani R, et al. Spectral imaging with scattered light: from early cancer detection to cell biology. IEEE J Sel Top Quantum Electron. 2012; 18:1073–1083. PMID: 23087592.

23. Perelman LT, Backman V, Wallace M, et al. Observation of periodic fine structure in reflectance from biological tissue: a new technique for measuring nuclear size distribution. Phys Rev Lett. 1998; 80:627–630.

24. Bedard N, Hagen N, Gao L, Tkaczyk TS. Image mapping spectrometry: calibration and characterization. Opt Eng. 2012; 51:pii: 111711.

25. Altelaar AF, Munoz J, Heck AJ. Next-generation proteomics: towards an integrative view of proteome dynamics. Nat Rev Genet. 2013; 14:35–48. PMID: 23207911.

26. Stunnenberg HG, Hubner NC. Genomics meets proteomics: identifying the culprits in disease. Hum Genet. 2013; 10. 18. Epub. DOI: 10.1007/s00439-013-1376-2.

27. Cramer R. Editorial for "advances in biological mass spectrometry and proteomics". Methods. 2011; 54:349–350. PMID: 21839395.

28. Filiou MD, Martins-de-Souza D, Guest PC, Bahn S, Turck CW. To label or not to label: applications of quantitative proteomics in neuroscience research. Proteomics. 2012; 12:736–747. PMID: 22247077.

29. Sjödin MO, Wetterhall M, Kultima K, Artemenko K. Comparative study of label and label-free techniques using shotgun proteomics for relative protein quantification. J Chromatogr B Analyt Technol Biomed Life Sci. 2013; 928:83–92.

30. Han NY, Choi W, Park JM, Kim EH, Lee H, Hahm KB. Label-free quantification for discovering novel biomarkers in the diagnosis and assessment of disease activity in inflammatory bowel disease. J Dig Dis. 2013; 14:166–174. PMID: 23320753.

31. Matsumura CY, Menezes de Oliveira B, Durbeej M, Marques MJ. Isobaric tagging-based quantification for proteomic analysis: a comparative study of spared and affected muscles from mice at the early phase of dystrophy. PLoS One. 2013; 8:e65831. PMID: 23823696.
32. Ko KH, Kwon CI, Park SH, et al. Application of matrix-assisted laser desorption/Ionization time-of-flight imaging mass spectrometry (MALDI-TOF IMS) for premalignant gastrointestinal lesions. Clin Endosc. 2013; 46:611–619. PMID: 24340253.

Fig. 1
(A) Animal model for Barrett esophagus (BE). Esophagojejunostomy was performed in Sprague Dawley rats to expose the esophagus to gastroduodenal contents. Histological examination showed the clear appearance of BE with partial changes of BE-associated adenocarcinoma (arrows). (B) cDNA microarray for biomarkers of BE. A 20,000 rat cDNA microarray (Macrogen) was probed using Cy3 and Cy5 labeling to identify the genes responsible for BE and BE-associated carcinoma with Scatchard plotting and bioinformatics analysis. The results are currently being validated.
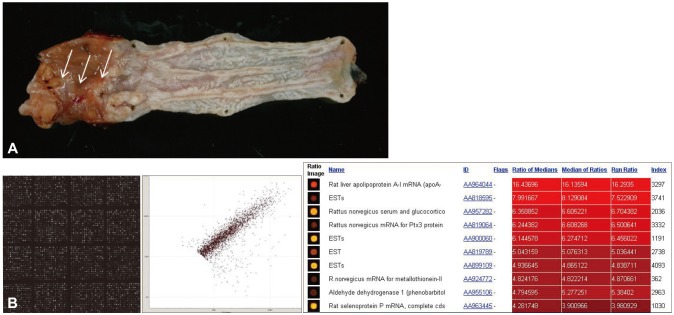
Fig. 2
Advances in molecular imaging technology for future medicine in gastroenterology, and cDNA microarray and imaging mass spectrometry (IMS). (A) Flow for IMS as exemplified in colitic cancer. (B) Label-free protein quantification scheme for either biomarker discovery or IMS. (C) Label-based protein quantification scheme using isobaric tags for relative and absolute quantification (iTRAQ) labeling. ITO, indium tin oxide; MALDI-TOF-IMS, matrix-assisted laser desorption/ionization time-of-flight imaging mass spectrometry; LC-MS/MS, liquid chromatography-tandem mass spectrometry; RT, chromatographic retention time; m/z, mass-to-charge ratio; PCA, principal components analysis.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download