A 17-year-old female visited our physical medicine and rehabilitation clinic complaining of toe walking with cognitive impairment and dysarthria. Her symptoms initiated several years back and progressed slowly; however, she had never had these symptoms evaluated by a medical practitioner. There were no remarkable findings in her genealogical chart; her elder sister had a normal development (
Fig. 1), and the patient's birth history was uneventful. On neurological examination, deep tendon reflexes were exaggerated on both the knees and ankles. The Modified Ashworth Scale of knee extension and flexion was grade 2 and 1+, respectively. The motor power of her legs was of fair grade with equinovarus deformities on both the feet, and there was no visible muscle atrophy. Electrodiagnostic studies, including nerve conduction, needle electromyography, and somatosensory and motor evoked potential studies, did not indicate any abnormality. She showed mild cognitive impairment; her Montreal Cognitive Assessment score was 21/30, and her receptive and vocabulary abilities were about 3–6 years lower than those appropriate for her age. The laboratory studies did not reveal any specific findings that could have suggested diseases presenting dysfunction of the central nervous system, such as Wilson's disease, autoimmune diseases, endocrine diseases, or metabolic diseases. Whole spine magnetic resonance imaging (MRI) presented no definite abnormalities; however, brain MRI showed thinning of the corpus callosum (
Fig. 2). HSP was suspected based on the clinical manifestations, sporadic spastic paraplegia (SPG), and no evidence of other primary etiology. SPG type 11 was strongly suspected as thinning of the corpus callosum is a common feature in SPG11; however, it is also found in other SPG subtypes. Therefore, sequencing of SPG4 (
SPAST) and SPG3A (
ATL1), which presents with relatively high frequency among individuals with HSP and
SPG11, was performed. Among the 40 coding exons of
SPG11, nine exons namely exons 2, 6, 7, 8, 30, 36, 37, 38, and 39 presented relatively high frequency of mutations, and the adjacent introns were analyzed by Sanger sequencing. Sequence analysis of
SPG11 in clinical practice is limited in Korea, since safety and effectiveness of the test has not yet been proved by National Evidence-based Healthcare Collaborating Agency. Therefore, genetic evaluation of
SPG11 is only available for the purpose of research and not clinical diagnosis. Based on these reasons, we selectively sequenced exons with a high frequency of
SPG11, rather than all exons and introns of
SPG11 gene. However, gene sequencing and chromosome analyses, including chromosome microarray, did not show any abnormal findings. Thereafter, she visited another clinic to further attempt to find a diagnosis for her disease. A neurologist from another center assumed her disease to be spinocerebellar ataxia (SCA). Fluorescent fragment length analysis of SCA type 1, 2, 3, 6, 7, 8, and 17 was performed; however, no mutations were detected. A few months later, she came to our center again for further detailed genetic studies. Clinically, HSP, especially SPG types 11 and 15, and SCA were suspected. However, DNA sequencing of exons with high incidence mutations did not reveal any matching results. Meanwhile, we were conscious about an NGS technology named CES that can analyze 4,813 genes and about 62,000 exomes associated with many genetic diseases in humans [
3]. The CES panel provides comprehensive coverage of HSP and SCA, including 50 of 67 known protein-encoding genes involved in HSP [
4] and 33 of 41 genes in SCA although some subtypes of SCA which are a result of trinucleotide repeat expansion could not be accurately detected [
5]. Genomic DNA was extracted from the peripheral blood of the patient and her parents and sister. The genomic DNA was enriched using the TruSight One Sequencing Panel, which includes 125,395 probes targeting a 12-Mb region spanning 4,813 genes, and was sequenced on the Illumina MiSeq platform [
3]. The size of probe is 80-mer, and it targets libraries of approximately 500 bp enriching 350–650 bases centered symmetrically on the midpoint of the probe. It means that, the panel provides coverage of exon-flanking regions, which is splice sites. Enrichment-ready fragmentation by tagmentation and PCR amplification involved 50 ng of input DNA. Pooled sample libraries were denatured and labeled by biotin-labeled probes specific to the targeted region for hybridization. The streptavidin beads were added and bound to the biotinylated probes. Biotinylated DNA fragments bound to the streptavidin-coated beads were magnetically pulled down from the solution, and the beads were removed. Captured targeted regions were loaded on MiSeq/NextSeq/Hiseq system for sequencing. On-instrument software automatically performed alignment and variant calling. Imported sequence data to the VariantStudio software was filtered and customized reporting for specified genes. CES revealed compound heterozygous variations on the patient's
SPG11 gene; a splice site aberration in intron 18 (NM_025137.3:c.3291+1G>T) and a nonsense variation in exon 16 (NM_025137.3:c.3036C>A, p.Try1012*). Each of the variations was reported as a pathogenic variant in the previous reports [
67]. The results also included other variants; however, they were not pathogenic but normal variants. Furthermore, Sanger sequencing revealed her parents as being heterozygous carriers for each mutation, and her sister as a carrier of heterozygous mutation (c.3036C>A/p.Try1012*) (
Table 1,
Fig. 3).
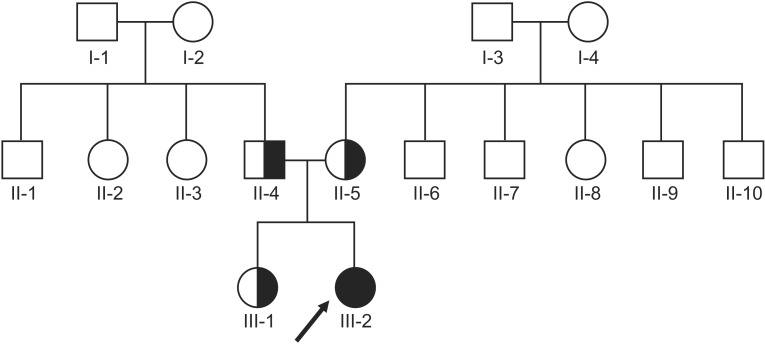 | Fig. 1Pedigree of the family of the 17-year-old female with spastic paraplegia. The black symbol represents the patient. The index I-3 died due to old age, and the index II-6 passed away at an early age due to an unknown cause.
|
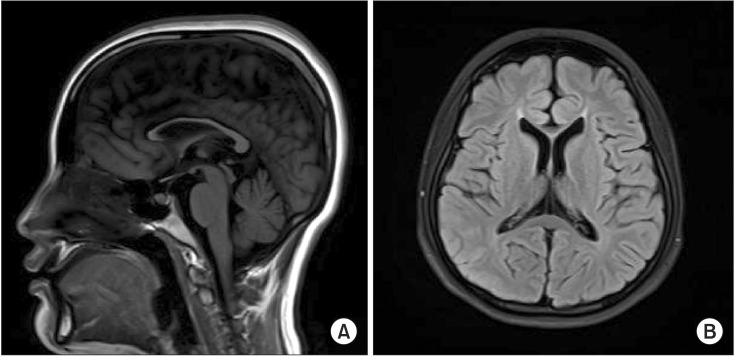 | Fig. 2Brain magnetic resonance imaging findings in the patient. (A) Sagittal T1-weighted image reveals a thin corpus callosum. (B) Axial T2-weighted image also shows a thin corpus callosum.
|
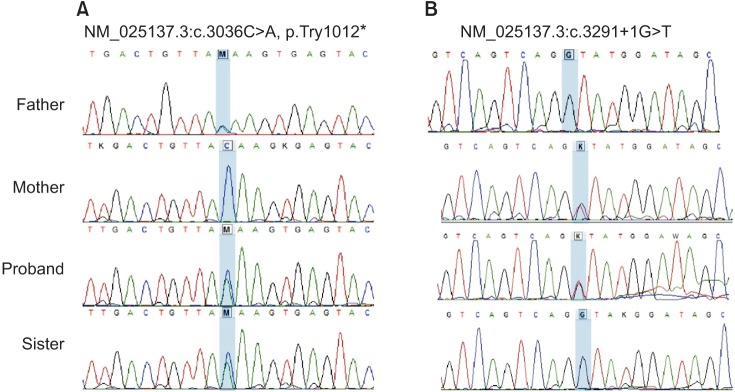 | Fig. 3DNA sequence chromatography for the patient and her family. (A) Heterozygous c.3036C>A mutations in the patient, her father and sister. (B) Heterozygous c.3291+1G>T mutations in the patient and her mother.
|
Table 1
Mutations on SPG11 of the patient and her family








 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download