1. Levey AS, Atkins R, Coresh J, Cohen EP, Collins AJ, Eckardt KU, et al. Chronic kidney disease as a global public health problem: approaches and initiatives - a position statement from Kidney Disease Improving Global Outcomes. Kidney Int. 2007; 72(3):247–259. PMID:
17568785.

2. Boron WF, Boulpaep EL. Medical Physiology E-book. Philadelphia, PA, USA: Elsevier Health Sciences;2016.
3. Tomasoni S, Benigni A. Gene therapy: how to target the kidney. Promises and pitfalls. Curr Gene Ther. 2004; 4(1):115–122. PMID:
15032618.

4. Hamar P, Song E, Kökény G, Chen A, Ouyang N, Lieberman J. Small interfering RNA targeting Fas protects mice against renal ischemia-reperfusion injury. Proc Natl Acad Sci U S A. 2004; 101(41):14883–14888. PMID:
15466709.

5. Takabatake Y, Isaka Y, Mizui M, Kawachi H, Takahara S, Imai E. Chemically modified siRNA prolonged RNA interference in renal disease. Biochem Biophys Res Commun. 2007; 363(2):432–437. PMID:
17880921.

6. Miyazawa H, Hirai K, Ookawara S, Ishibashi K, Morishita Y. Nano-sized carriers in gene therapy for renal fibrosis
in vivo
. Nano Rev Exp. 2017; 8(1):1331099. PMID:
30410705.
7. Lai LW, Moeckel GW, Lien YH. Kidney-targeted liposome-mediated gene transfer in mice. Gene Ther. 1997; 4(5):426–431. PMID:
9274719.

8. Ito K, Chen J, Asano T, Vaughan ED Jr, Poppas DP, Hayakawa M, et al. Liposome-mediated gene therapy in the kidney. Hum Cell. 2004; 17(1):17–28. PMID:
15369133.

9. Morishita Y, Imai T, Yoshizawa H, Watanabe M, Ishibashi K, Muto S, et al. Delivery of microRNA-146a with polyethylenimine nanoparticles inhibits renal fibrosis in vivo. Int J Nanomedicine. 2015; 10:3475–3488. PMID:
25999712.

10. Xia Z, Abe K, Furusu A, Miyazaki M, Obata Y, Tabata Y, et al. Suppression of renal tubulointerstitial fibrosis by small interfering RNA targeting heat shock protein 47. Am J Nephrol. 2008; 28(1):34–46. PMID:
17890856.

11. Aoyama T, Yamamoto S, Kanematsu A, Ogawa O, Tabata Y. Local delivery of matrix metalloproteinase gene prevents the onset of renal sclerosis in streptozotocin-induced diabetic mice. Tissue Eng. 2003; 9(6):1289–1299. PMID:
14674437.

12. Pillarisetti S, Uthaman S, Huh KM, Koh YS, Lee S, Park IK. Multimodal composite iron oxide nanoparticles for biomedical applications. Tissue Eng Regen Med. 2019; 16(5):451–465. PMID:
31624701.

13. Imai E, Takabatake Y, Mizui M, Isaka Y. Gene therapy in renal diseases. Kidney Int. 2004; 65(5):1551–1555. PMID:
15086890.

14. Kim CS, Mathew AP, Uthaman S, Moon MJ, Bae EH, Kim SW, et al. Glycol chitosan-based renal docking biopolymeric nanomicelles for site-specific delivery of the immunosuppressant. Carbohydr Polym. 2020; 241:116255. PMID:
32507186.

15. Lee JN, Chun SY, Ha YS, Choi KH, Yoon GS, Kim HT, et al. Target molecule expression profiles in metastatic renal cell carcinoma: development of individual targeted therapy. Tissue Eng Regen Med. 2016; 13(4):416–427. PMID:
30603423.

16. Singh RK, Kim HW. Inorganic nanobiomaterial drug carriers for medicine. Tissue Eng Regen Med. 2013; 10(6):296–309.

17. Longmire MR, Ogawa M, Choyke PL, Kobayashi H. Biologically optimized nanosized molecules and particles: more than just size. Bioconjug Chem. 2011; 22(6):993–1000. PMID:
21513351.

18. Mohanty A, Uthaman S, Park IK. Utilization of polymer-lipid hybrid nanoparticles for targeted anti-cancer therapy. Molecules. 2020; 25(19):4377.

19. Choi H, Choi Y, Yim HY, Mirzaaghasi A, Yoo JK, Choi C. Biodistribution of exosomes and engineering strategies for targeted delivery of therapeutic exosomes. Tissue Eng Regen Med. 2021; 18(4):499–511. PMID:
34260047.

20. Huang Y, Wang J, Jiang K, Chung EJ. Improving kidney targeting: The influence of nanoparticle physicochemical properties on kidney interactions. J Control Release. 2021; 334:127–137. PMID:
33892054.

21. Yang C, Nilsson L, Cheema MU, Wang Y, Frøkiær J, Gao S, et al. Chitosan/siRNA nanoparticles targeting cyclooxygenase type 2 attenuate unilateral ureteral obstruction-induced kidney injury in mice. Theranostics. 2015; 5(2):110–123. PMID:
25553102.

22. Gao S, Hein S, Dagnæs-Hansen F, Weyer K, Yang C, Nielsen R, et al. Megalin-mediated specific uptake of chitosan/siRNA nanoparticles in mouse kidney proximal tubule epithelial cells enables AQP1 gene silencing. Theranostics. 2014; 4(10):1039–1051. PMID:
25157280.

23. Suh SH, Mathew AP, Choi HS, Vasukutty A, Kim CS, Kim IJ, et al. Kidney-accumulating olmesartan-loaded nanomicelles ameliorate the organ damage in a murine model of Alport syndrome. Int J Pharm. 2021; 600:120497. PMID:
33753165.

24. Kim CS, Mathew AP, Vasukutty A, Uthaman S, Joo SY, Bae EH, et al. Glycol chitosan-based tacrolimus-loaded nanomicelle therapy ameliorates lupus nephritis. J Nanobiotechnology. 2021; 19(1):109. PMID:
33865397.

25. Vansthertem D, Gossiaux A, Declèves AE, Caron N, Nonclercq D, Legrand A, et al. Expression of nestin, vimentin, and NCAM by renal interstitial cells after ischemic tubular injury. J Biomed Biotechnol. 2010; 2010:193259. PMID:
20617137.

26. Holthöfer H, Miettinen A, Lehto VP, Lehtonen E, Virtanen I. Expression of vimentin and cytokeratin types of intermediate filament proteins in developing and adult human kidneys. Lab Invest. 1984; 50(5):552–559. PMID:
6201675.
27. Beham A, Ratschek M, Zatloukal K, Schmid C, Denk H. Distribution of cytokeratins, vimentin and desmoplakins in normal renal tissue, renal cell carcinomas and oncocytoma as revealed by immunofluorescence microscopy. Virchows Arch A Pathol Anat Histopathol. 1992; 421(3):209–215. PMID:
1384221.

28. Skinnider BF, Folpe AL, Hennigar RA, Lim SD, Cohen C, Tamboli P, et al. Distribution of cytokeratins and vimentin in adult renal neoplasms and normal renal tissue: potential utility of a cytokeratin antibody panel in the differential diagnosis of renal tumors. Am J Surg Pathol. 2005; 29(6):747–754. PMID:
15897741.
29. Ise H, Kobayashi S, Goto M, Sato T, Kawakubo M, Takahashi M, et al. Vimentin and desmin possess GlcNAc-binding lectin-like properties on cell surfaces. Glycobiology. 2010; 20(7):843–864. PMID:
20332081.

30. Aso S, Ise H, Takahashi M, Kobayashi S, Morimoto H, Izawa A, et al. Effective uptake of N-acetylglucosamine-conjugated liposomes by cardiomyocytes in vitro. J Control Release. 2007; 122(2):189–198. PMID:
17681632.

31. Singh B, Maharjan S, Kim YK, Jiang T, Islam MA, Kang SK, et al. Targeted gene delivery via N-acetylglucosamine receptor mediated endocytosis. J Nanosci Nanotechnol. 2014; 14(11):8356–8364. PMID:
25958528.

32. Kim SJ, Ise H, Goto M, Komura K, Cho CS, Akaike T. Gene delivery system based on highly specific recognition of surface-vimentin with N-acetylglucosamine immobilized polyethylenimine. Biomaterials. 2011; 32(13):3471–3480. PMID:
21329974.

33. Lungwitz U, Breunig M, Blunk T, Göpferich A. Polyethylenimine-based non-viral gene delivery systems. Eur J Pharm Biopharm. 2005; 60(2):247–266. PMID:
15939236.

34. Fischer D, Bieber T, Li Y, Elsässer HP, Kissel T. A novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: effect of molecular weight on transfection efficiency and cytotoxicity. Pharm Res. 1999; 16(8):1273–1279. PMID:
10468031.
35. Forrest ML, Koerber JT, Pack DW. A degradable polyethylenimine derivative with low toxicity for highly efficient gene delivery. Bioconjug Chem. 2003; 14(5):934–940. PMID:
13129396.

36. Ahn CH, Chae SY, Bae YH, Kim SW. Biodegradable poly(ethylenimine) for plasmid DNA delivery. J Control Release. 2002; 80(1-3):273–282. PMID:
11943404.

37. Maiti KK, Lee WS, Takeuchi T, Watkins C, Fretz M, Kim DC, et al. Guanidine-containing molecular transporters: sorbitol-based transporters show high intracellular selectivity toward mitochondria. Angew Chem Int Ed Engl. 2007; 46(31):5880–5884. PMID:
17607670.

38. Shaikh SF, Mane RS, Min BK, Hwang YJ, Joo O. D-sorbitol-induced phase control of TiO 2 nanoparticles and its application for dye-sensitized solar cells. Sci Rep. 2016; 6(1):1–10. PMID:
28442746.

39. Islam MA, Yun CH, Choi YJ, Shin JY, Arote R, Jiang HL, et al. Accelerated gene transfer through a polysorbitol-based transporter mechanism. Biomaterials. 2011; 32(36):9908–9924. PMID:
21959011.

40. Islam MA, Shin JY, Firdous J, Park TE, Choi YJ, Cho MH, et al. The role of osmotic polysorbitol-based transporter in RNAi silencing via caveolae-mediated endocytosis and COX-2 expression. Biomaterials. 2012; 33(34):8868–8880. PMID:
22975426.

41. Islam MA, Shin JY, Yun CH, Cho CS, Seo HW, Chae C, et al. The effect of RNAi silencing of p62 using an osmotic polysorbitol transporter on autophagy and tumorigenesis in lungs of K-rasLA1 mice. Biomaterials. 2014; 35(5):1584–1596. PMID:
24269155.

42. Cho WY, Hong SH, Singh B, Islam MA, Lee S, Lee AY, et al. Suppression of tumor growth in lung cancer xenograft model mice by poly(sorbitol-co-PEI)-mediated delivery of osteopontin siRNA. Eur J Pharm Biopharm. 2015; 94:450–462. PMID:
26141346.

43. Nguyen KC, Muthiah M, Islam MA, Kalash RS, Cho CS, Park H, et al. Selective transfection with osmotically active sorbitol modified PEI nanoparticles for enhanced anti-cancer gene therapy. Colloids Surf B Biointerfaces. 2014; 119:126–136. PMID:
24880989.

44. Steenhard BM, Vanacore R, Friedman D, Zelenchuk A, Stroganova L, Isom K, et al. Upregulated expression of integrin α1 in mesangial cells and integrin α3 and vimentin in podocytes of Col4a3-null (Alport) mice. PLoS One. 2012; 7(12):e50745. PMID:
23236390.

45. Kim YK, Singh B, Jiang HL, Park TE, Jiang T, Park IK, et al. N-Acetylglucosamine-conjugated block copolymer consisting of poly(ethylene oxide) and cationic polyaspartamide as a gene carrier for targeting vimentin-expressing cells. Eur J Pharm Sci. 2014; 51:165–172. PMID:
24075972.

46. Van Nguyen TT, Vu NB, Van Pham P. Mesenchymal stem cell transplantation for ischemic diseases: mechanisms and challenges. Tissue Eng Regen Med. 2021; 18(4):587–611. PMID:
33884577.

47. Islam MA, Kim S, Firdous J, Lee AY, Hong SH, Seo MK, et al. A high affinity kidney targeting by chitobionic acid-conjugated polysorbitol gene transporter alleviates unilateral ureteral obstruction in rats. Biomaterials. 2016; 102:43–57. PMID:
27318934.

48. Kaifu R, Goldstein IJ. N,N′-Diacetylchitobionic N-alkylamides: synthesis, and interaction with two 2-acetamido-2-deoxy-D-glucose-binding lectins. Carbohydr Res. 1981; 96(2):241–247. PMID:
6895483.

49. Luu QP, Shin JY, Kim YK, Islam MA, Kang SK, Cho MH, et al. High gene transfer by the osmotic polysorbitol-mediated transporter through the selective caveolae endocytic pathway. Mol Pharm. 2012; 9(8):2206–2218. PMID:
22708896.

50. Inada M, Izawa G, Kobayashi W, Ozawa M. 293 cells express both epithelial as well as mesenchymal cell adhesion molecules. Int J Mol Med. 2016; 37(6):1521–1527. PMID:
27121032.

51. Ooi A, Wong A, Esau L, Lemtiri-Chlieh F, Gehring C. A guide to transient expression of membrane proteins in HEK-293 cells for functional characterization. Front Physiol. 2016; 7:300. PMID:
27486406.

52. Hou S, Ziebacz N, Wieczorek SA, Kalwarczyk E, Sashuk V, Kalwarczyk T, et al. Formation and structure of PEI/DNA complexes: quantitative analysis. Soft Matter. 2011; 7(15):6967–6972.

53. Hendrix MJ, Seftor EA, Seftor RE, Trevor KT. Experimental co-expression of vimentin and keratin intermediate filaments in human breast cancer cells results in phenotypic interconversion and increased invasive behavior. Am J Pathol. 1997; 150(2):483–495. PMID:
9033265.
54. Kusaba T, Lalli M, Kramann R, Kobayashi A, Humphreys BD. Differentiated kidney epithelial cells repair injured proximal tubule. Proc Natl Acad Sci U S A. 2014; 111(4):1527–1532. PMID:
24127583.

55. Ramos I, Stamatakis K, Oeste CL, Pérez-Sala D. Vimentin as a multifaceted player and potential therapeutic target in viral infections. Int J Mol Sci. 2020; 21(13):4675.

56. Wang Z, Divanyan A, Jourd’heuil FL, Goldman RD, Ridge KM, Jourd’heuil D, et al. Vimentin expression is required for the development of EMT-related renal fibrosis following unilateral ureteral obstruction in mice. Am J Physiol Renal Physiol. 2018; 315(4):F769–F780. PMID:
29631355.



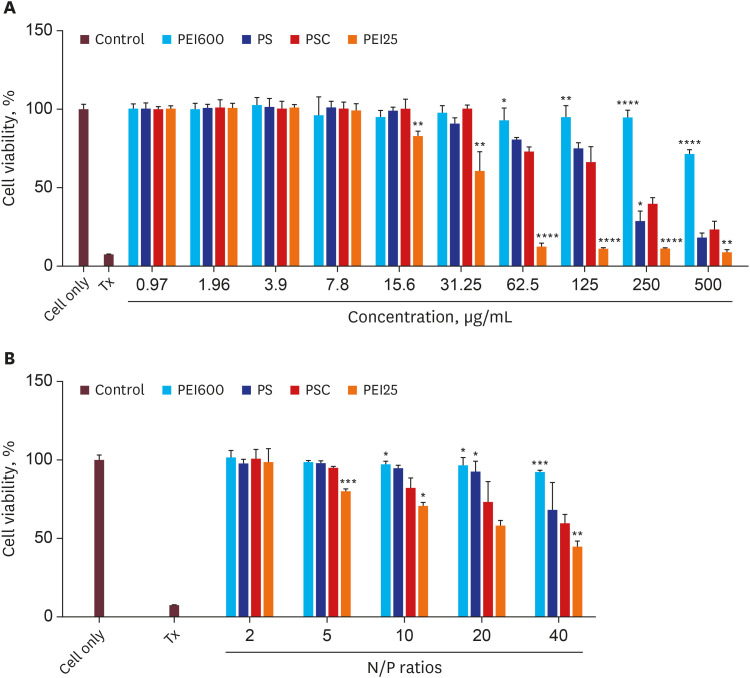




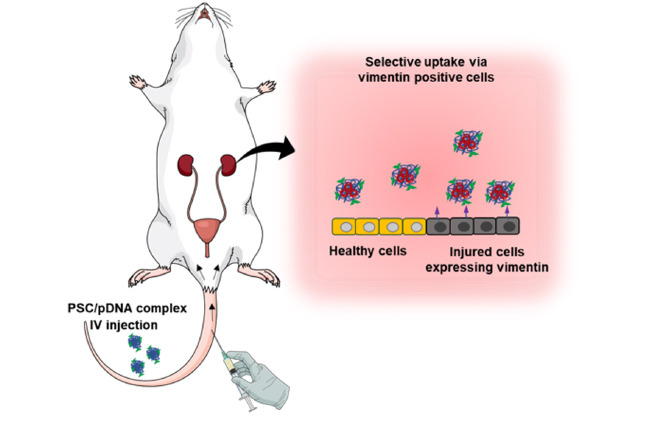
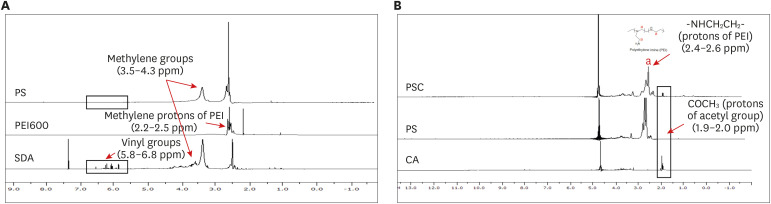
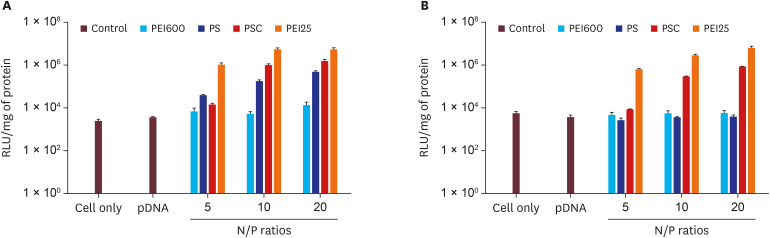
 PDF
PDF Citation
Citation Print
Print






 XML Download
XML Download