INTRODUCTION
The coronavirus disease 2019 (COVID-19) pandemic is an ongoing global health crisis caused by a newly discovered coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
1 The COVID-19 pandemic began in December 2019 in Wuhan City, China, and has lasted for more than a year worldwide.
2 Clinical presentation of COVID-19 ranges from asymptomatic cases and mild illness to critical conditions featuring acute respiratory distress syndrome.
3 In Korea, patients less than 10 years and over 70 years are less frequently infected with COVID-19, and those aged between 20–59 years are more commonly infected.
4 COVID-19 disease severity is associated with increased age.
5 Influenza virus (Flu) and respiratory syncytial virus (RSV) are common respiratory pathogens that cause seasonal epidemics.
67 Influenza infection tend to be less severe, typically resulting in uncomplicated upper respiratory tract illness, but, in rare cases, influenza infection can induce a complicated disease with severe viral pneumonia.
8 Children are most likely to be infected with influenza, and people aged 65 years and older are least likely to become ill from influenza.
9 RSV is recognized as the most common cause of bronchiolitis and pneumonia in children younger than 1 year. RSV infection can produce common cold-like symptoms but may be severe in infants and young children. In older adults, RSV usually causes no symptoms or upper respiratory infection but may lead to severe lower respiratory infection.
10
SARS-CoV-2, influenza, and RSV infections should be managed in different ways. COVID-19 management requires strict quarantine and isolation strategies that are not applied to influenza and RSV infection management. Among these three infective diseases, influenza is the only effectively treatable condition; influenza can be treated with the drug, oseltamivir (Tamiflu).
Since the clinical presentations of SARS-CoV-2, influenza, and RSV infections have overlapping symptoms, differential diagnosis of these diseases is challenging. Specifically, rapid diagnosis is needed in elderly patients because SARS-CoV-2, influenza, and RSV infections result in substantial morbidity and mortality in these patients.
611 In the COVID-19 pandemic era, clinicians may encounter a co-infection of SARS-CoV-2 with influenza or RSV.
121314 According to a
Nature survey, many researchers expect SARS-CoV-2 to become endemic.
15 In the future, SARS-CoV-2, influenza, and RSV are expected to be the most common respiratory viruses infected in humans. Therefore, multiplex detection of SARS-CoV-2, influenza, and RSV will be essential and commonly used.
Molecular testing is the gold standard for the detection of many viruses, and real-time reverse transcription polymerase chain reaction (rRT-PCR) is most widely used molecular method for qualitative diagnostic testing.
16 Numerous rRT-PCR kits that solely detect SARS-CoV-2 have been developed and commercialized worldwide, including in Korea.
171819 Seegene has the highest market share in the SARS-CoV-2 single molecular diagnostic kit market in Korea, followed by KogeneBiotech and SD BioSensor. The United States Food and Drug Administration approved the use of SARS-CoV-2 molecular diagnostic kits produced by these three companies under Emergency Use Authorization in April 2020.
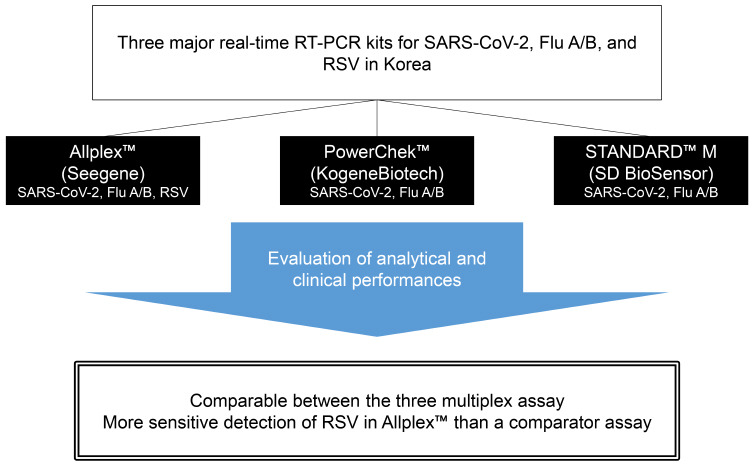
20 Recently, these three manufacturers have developed molecular diagnostic kits for concurrent detection of SARS-CoV-2, influenza A/B, and RSV. The kits produced by Seegene and KogeneBiotech were approved for use by the Ministry of Food and Drug Safety in Korea in January 2021 and November 2020, respectively, although the kit produced by SD BioSensor has not yet been approved (last reviewed on October 20, 2021). In this study, we evaluated the analytical and clinical performance of these three multiplex rRT-PCR assays for simultaneous detection of SARS-CoV-2, influenza A/B, and RSV in nasopharyngeal swabs (NPS).
METHODS
Three molecular diagnostic kits for simultaneous detection of SARS-CoV-2 and influenza A/B and/or RSV
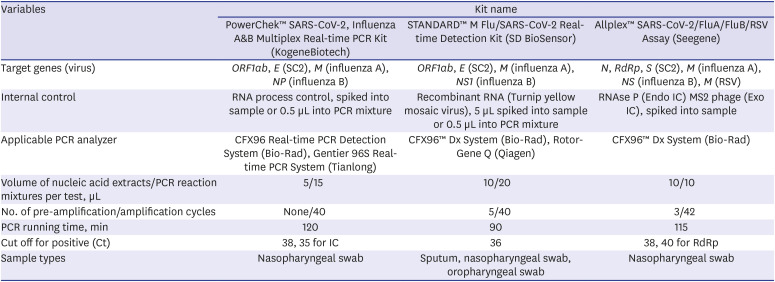
PowerChek™ SARS-CoV-2, Influenza A&B Multiplex Real-time PCR Kit (PowerChek; KogeneBiotech, Seoul, Korea) and STANDARD™ M Flu/SARS-CoV-2 Real-time Detection Kit (STANDARD M; SD BioSensor, Osong, Korea) are multiplex kits for the detection of SARS-CoV-2 and influenza A/B. Allplex™ SARS-CoV-2/FluA/FluB/RSV Assay (Allplex; Seegene, Seoul, Korea) is a multiplex kit for the detection of SARS-CoV-2, influenza A/B, and RSV.
To detect SARS-CoV-2, PowerChek™ SARS-CoV-2, Influenza A&B Multiplex Real-time PCR Kit (PowerChek; KogeneBiotech), and STANDARD™ M Flu/SARS-CoV-2 Real-time Detection Kit (STANDARD M; SD BioSensor) target the
envelope (E) and
open reading frame 1ab (ORF1ab) coding regions of the SARS-CoV-2 genome, whereas Allplex™ SARS-CoV-2/FluA/FluB/RSV Assay (Allplex; Seegene, Seoul, Korea) targets the
nucleocapsid (N),
RNA-dependent RNA polymerase (RdRp), and
spike (S) coding regions. According to the World Health Organization guideline,
21 rRT-PCR results are interpreted as positive when all target genes are detected. If no target genes are detected, the test is interpreted as negative. The cases where the target genes are partially detected are considered inconclusive. For the detection of influenza A, PowerChek, STANDARD M, and Allplex target the
matrix (M) genes. For the detection of influenza B, PowerChek, STANDARD M, and Allplex target the
nucleoprotein (NP),
nonstructural (NS) 1, and
NS genes, respectively. Allplex targets the
matrix (M) gene for the detection of RSV. All kits are based on one-step rRT-PCR in one tube. Specifications of the three assays are described in
Table 1.
Table 1
Specification of three commercial real-time RT-PCR kits for the multiplex detection of SARS-CoV-2, influenza virus A/B, and/or RSV
|
Variables |
Kit name |
|
PowerChek™ SARS-CoV-2, Influenza A&B Multiplex Real-time PCR Kit (KogeneBiotech) |
STANDARD™ M Flu/SARS-CoV-2 Real-time Detection Kit (SD BioSensor) |
Allplex™ SARS-CoV-2/FluA/FluB/RSV Assay (Seegene) |
|
Target genes (virus) |
ORF1ab, E (SC2), M (influenza A), NP (influenza B) |
ORF1ab, E (SC2), M (influenza A), NS1 (influenza B) |
N, RdRp, S (SC2), M (influenza A), NS (influenza B), M (RSV) |
|
Internal control |
RNA process control, spiked into sample or 0.5 μL into PCR mixture |
Recombinant RNA (Turnip yellow mosaic virus), 5 μL spiked into sample or 0.5 μL into PCR mixture |
RNAse P (Endo IC) MS2 phage (Exo IC), spiked into sample |
|
Applicable PCR analyzer |
CFX96 Real-time PCR Detection System (Bio-Rad), Gentier 96S Real-time PCR System (Tianlong) |
CFX96™ Dx System (Bio-Rad), Rotor-Gene Q (Qiagen) |
CFX96™ Dx System (Bio-Rad) |
|
Volume of nucleic acid extracts/PCR reaction mixtures per test, μL |
5/15 |
10/20 |
10/10 |
|
No. of pre-amplification/amplification cycles |
None/40 |
5/40 |
3/42 |
|
PCR running time, min |
120 |
90 |
115 |
|
Cut off for positive (Ct) |
38, 35 for IC |
36 |
38, 40 for RdRp |
|
Sample types |
Nasopharyngeal swab |
Sputum, nasopharyngeal swab, oropharyngeal swab |
Nasopharyngeal swab |

Specimen collection
We selected 103 SARS-CoV-2 positive and 201 SARS-CoV-2 negative specimens that were collected from September 2020 to November 2020 and reported by STANDARD™ M new Coronavirus (nCoV) Real-Time Detection kit (STANDARD M nCoV; SD BioSensor). The specimens were nasopharyngeal and/or oropharyngeal swabs from patients. Aliquots of samples were stored at −80°C. We retrieved the aliquots and thawed them for testing using the three assays and STANDARD M nCoV. Re-testing using STANDARD M nCoV revealed that the results of six SARS-CoV-2 positive samples were changed from positive to inconclusive or negative, and the results of the remaining 97 SARS-CoV-2 positive samples were unchanged. We excluded the six specimens and finally included 97 SARS-CoV-2-positive and 201 SARS-CoV-2-negative specimens. All 97 SARS-CoV-2-positive samples were acquired from confirmed cases.
Four hundred and three respiratory virus-positive NPS samples tested by AdvanSure™ respiratory viruses (RV) real-time RT-PCR assay (AdvanSure; LG Life Sciences, Seoul, Korea) were collected from December 2017 to November 2020. A substantial portion of the respiratory virus-positive NPS samples tested by the AdvanSure diagnostic kit was acquired before the COVID-19 pandemic. Aliquots of samples were stored at −80°C. We retrieved the aliquots and thawed them for testing using the three assays. When the results of the three assays were different from the previously reported results of the AdvanSure kit, the corresponding aliquots were tested with the AdvanSure kit again. We used re-tested AdvanSure results if tested again; if not, the previously reported results were used for analysis. All specimens harbored one or more respiratory viruses including 71 influenza A-positive, 50 influenza B-positive, 78 RSV-positive, and 207 other respiratory virus-positive samples.
Sample processing and PCR
A 200 μL volume of the patient specimen was extracted using the EMAG system (bioMérieux, Marcy l'Etoile, France) according to the manufacturer’s instructions, with a nucleic acid elution volume of 50 μL. All real-time PCR analyses were performed using the CFX96 Real-Time Detection System (Bio-Rad, Hercules, CA, USA). PCR reactions were performed in a total volume of 20 μL (15 μL PCR reaction mixture and 5 μL template RNA), a total volume of 30 μL (20 μL PCR reaction mixture and 10 μL template RNA), and a total volume of 20 μL (9 μL PCR reaction mixture and 11 μL template RNA) using the PowerChek, STANDARD M, and Allplex kits, respectively.
Analytical performance: limit of detection (LoD) and cross-reactivity
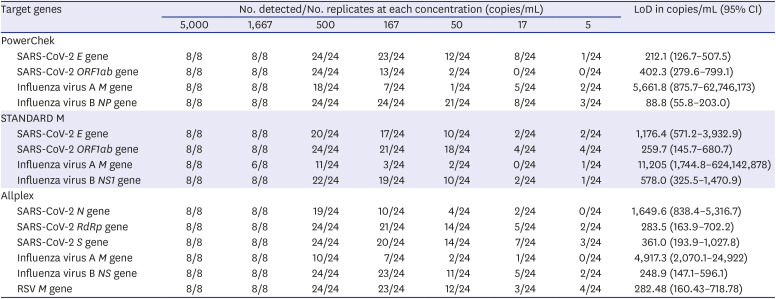
The LoD was determined using the AccuPlex™ SARS-CoV-2, Flu A/B, and RSV reference material kits (SeraCare, Milford, MA, USA). AccuPlex virus products contained non-replicative recombinant viruses including full genomes of SARS-CoV-2 and influenza A/B and partial genome of RSV (NC_001803, 1..4380; 8460..15191). We obtained seven concentrations (5,000, 1,667, 500, 167, 50, 17, and 5 copies/mL) by serial dilution from reference non-replicative recombinant viruses. The first two concentrations (5,000 and 1,667 copies/mL) were tested with eight replicates each. The other concentrations were tested with 24 replicates each. LoD was defined as the input copy number per 1 mL with a 95% probability of a positive PCR and was calculated by probit regression analysis using International Business Machines (IBM) Statistical Package for the Social Sciences software package (version 25; IBM, Armonk, NY, USA) with 95% confidence intervals (CIs).
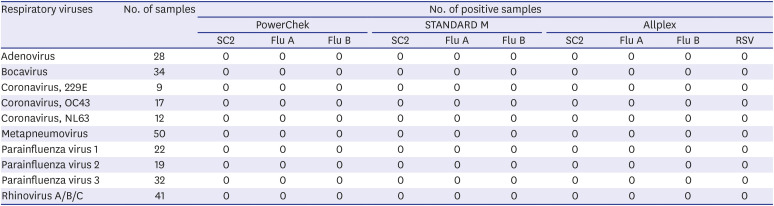
Cross-reactivity was verified using the 403 clinical samples containing typical respiratory viruses. Cross-reactivity was defined as the ability to generate a false-positive in similar viruses during RT-PCR analysis. Reference results were acquired using the AdvanSure diagnostic kits. Among the 403 respiratory virus-positive NPS samples, 47 samples tested positive for multiple respiratory pathogens. Since all three diagnostic kits were not regarded as cross-reactive to the 47 samples, each sample was regarded as multiple samples harboring different viruses.
Evaluation of the clinical performance of the three molecular diagnostic kits
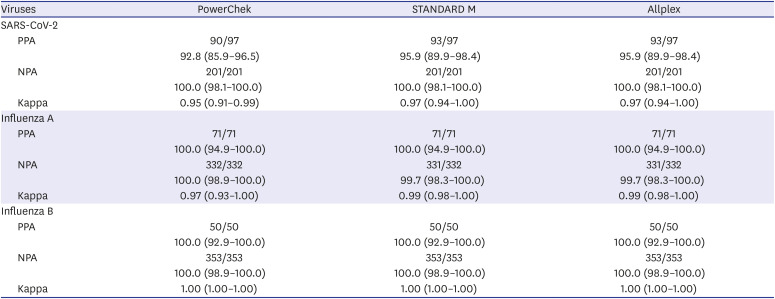
Reference results for positive and negative agreements were defined as follows: With regards to SARS-CoV-2, positive samples were defined as the sum of positive samples for all three kits and confirmed cases with discrepant results between three kits. Negative samples were defined as samples that tested negative for all three kits. In terms of influenza A/B, positive or negative samples were defined when two or three kits showed positive or negative results, respectively. When one kit produced discrepant results with the other two kits, the result of this kit was regarded as false.
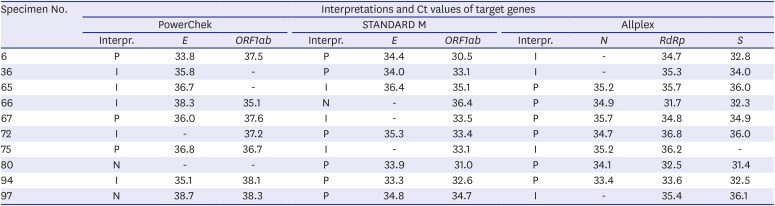
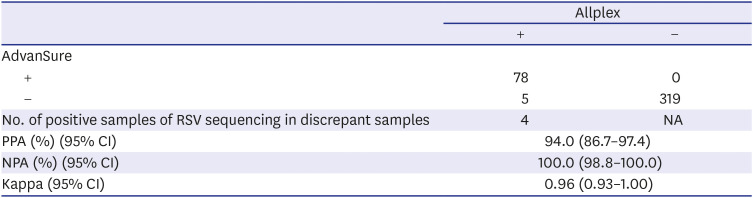
The RSV results of the Allplex diagnostic kit were compared with those of the AdvanSure comparator kit. The discordant results between the three kits and the AdvanSure comparator kit might be due to differences in sensitivity. Therefore, we estimated LoDs for influenza A/B and RSV in AdvanSure using the AccuPlex™ SARS-CoV-2, Flu A/B, and RSV reference material kits (SeraCare). RT-PCR and gene sequencing to identify RSV were performed on samples that produced discrepant results between Allplex and AdvanSure.
Positive percent agreements (PPAs) and negative percent agreements (NPAs) with 95% CIs were calculated using GraphPad Prism 7 (GraphPad, San Diego, CA, USA). Cohen’s kappa values with 95% CIs were calculated using GraphPad QuickCalcs (
https://www.graphpad.com/quickcalcs/kappa1/) (GraphPad).
Ethics statement
This study was approved by the Institutional Review Board of the Seoul National University Boramae Medical Center and the requirement for informed consent was waived (IRB No. 20-2020-262).
DISCUSSION
In the evaluation of analytical sensitivity, the LoD is dependent on reference material kits, extraction methods, PCR machines, and other experimental conditions. Therefore, the LoDs reported in our study may not correspond with practical LoDs in the real world. In the evaluation of cross-reactivity of the three assays, all three kits did not present cross-reactivity with common respiratory viruses. Especially, for SARS-CoV-2, three assays showed no reactive with coronavirus 229E, OC43, and NL63.
All three kits had 100% NPA in SARS-CoV-2 results, but PPAs in SARS-CoV-2 results ranged from 92.8% to 95.9%; among 97 SARS-CoV-2 positive samples, 10 (10.3%) produced discrepant results among the three assays. All three kits had 100% PPA in influenza A/B results. In contrast to 100% NPA in influenza B results in all three kits, NPAs in influenza A results ranged from 99.7% to 100.0% due to two false influenza A-positive reactions. According to the LoD testing of AdvanSure and Allplex kits and RSV sequencing results, Allplex detected RSV more sensitively than AdvanSure.
Molecular diagnostic kits for detecting SARS-CoV-2 have shifted from single SARS-CoV-2 detection to multi-detection of SARS-CoV-2 and other common respiratory pathogens. Nörz et al.
23 adapted a laboratory-developed multiplex RT-PCR assay for the simultaneous detection of SARS-CoV-2 and influenza A/B on a fully automated high-throughput system and demonstrated analytical performance comparable with that of currently available commercial tests. Sensitivities of SARS-CoV-2, influenza A, and influenza B in the multiplex assay were 98.1%, 97.67%, and 100%, respectively.
23 Xpert Xpress SARS-CoV-2/Flu-RSV (Xpert 4-in-1 assay) was launched with Emergency Use Authorization of the United States Food and Drug Administration in September 2020 and demonstrated a performance highly comparable with those of Xpert SARS-CoV-2 and Xpert Flu/RSV assays.
24 PPAs and NPAs of SARS-CoV-2, influenza A, influenza B, and RSV in Xpert -in-1 assay were 98.48%/100%, 100%/100%, 100%/99.54%, and 100%/100%, respectively.
24
A case of co-infection of SARS-CoV-2 and other respiratory viruses was absent in the present study. SARS-CoV-2-positive specimens were collected between September 2020 and November 2020. In Korea, RSV and influenza outbreak seasons classically begin in autumn and winter, respectively.
25 Although the collection period overlapped with RSV outbreak season, due to COVID-19-related quarantine and improved hygiene, transmission rates of influenza and RSV were lower.
26 A US research team reported that 20.7% of specimens positive for SARS-CoV-2 were positive for one or more additional pathogens, compared with 26.7% negative for SARS-CoV-2.
27 The most commonly co-infected pathogens were rhinovirus/enterovirus followed by RSV and non-SARS-CoV-2 Coronaviridae.
27 A single-centered retrospective study was performed in Wuhan, China, between January 12 and February 21, 2020 during the initial COVID-19 outbreak.
28 Unexpectedly, among COVID-19 patients, only 42.7% (131/307) of patients were singly positive for SARS-CoV-2; 57.3% (176/307) of COVID-19 patients were also positive for influenza viruses including influenza A (49.8%) and influenza B (7.5%).
28 Another single-centered retrospective study conducted in Lukou District, China showed that 14.1% (11/78) of COVID-19 patients were co-infected with other respiratory pathogens including RSV (3/11, 36.3%) and influenza B (1/11, 9.1%).
14 In contrast, a Korean research team demonstrated co-infection cases of SARS-CoV-2 and influenza virus or RSV were absent.
29 Using the Allplex RV-Essential Assay (Seegene) detecting seven respiratory viruses including adenovirus, influenza A virus, influenza B virus, metapneumovirus, parainfluenza virus, RSV, and human rhinovirus, the co-infection rate in COVID-19 patients was 2.2%.
29 The most frequently detected virus was adenovirus, followed by human rhinovirus and metapneumovirus.
29 However, comprehensive PCR testing to detect respiratory viruses other than influenza virus and RSV was not performed in SARS-CoV-2-positive specimens in the present study.
PowerChek, STANDARD M, and Allplex kits each specified different cycle conditions for rRT-PCR. PowerChek did not require any pre-amplification cycles during PCR. In the PCR protocol of STANDARD M, five pre-amplification cycles preceded the amplification cycles, and the time and temperature conditions of both cycles were the same. Allplex had three pre-amplification cycles with different time and temperature conditions from those of the amplification cycles. Volume of template RNA per test and the number of pre-amplification cycles and amplification cycles of the three multi-target kits were compared with corresponding single SARS-CoV-2 detection kits: PowerChek 2019-nCoV Real-time PCR (Kogene Biotech), STANDARD M nCoV Real-Time Detection (SD BioSensor), and Allplex 2019-nCoV (Seegene).
17 KogeneBiotech and SD BioSensor had the same volume of template RNA per test and number of pre-amplification and amplification cycles in each pair of single- and multi-target assays. However, the multi-target assay of Allplex required 10 μL of template RNA compared with 8 μL in the single-target assay. Allplex single-target assay performed 45 amplification cycles without pre-amplification but the multi-target assay executed 3 pre-amplification and 42 amplification cycles. Allplex multi-target assay was thought to be designed to increase sensitivity than the single-target assay by increasing the volume of template RNA.
This study had some limitations. First, as previously mentioned, the evaluation of the LoD of the kits may not have reflected the practical analytical sensitivity because the measured LoD may have been affected by extraction method, type of PCR machine, etc. Second, for SARS-CoV-2, cross-reactivity test with other coronaviruses except 229E, OC43, and NL63 was not performed. Third, although the AdvanSure kit was treated as a comparator kit in this study, a true reference method was absent. Indeed, the AdvanSure kit was less sensitive for RSV compared with the Allplex kit. Finally, because multiplex PCR used to detect respiratory viruses other than influenza A/B and RSV was not performed in SARS-CoV-2-positive samples, co-infection rates of other respiratory viruses and SARS-CoV-2 could not be estimated.
In conclusion, the overall performance of these three multiplex rRT-PCR assays for concurrent detection of SARS-CoV-2, influenza A/B, and RSV was comparable. In the COVID-19 pandemic era, all three kits are suitable for the prompt differential diagnosis of COVID-19, influenza, and RSV infection in patients with respiratory symptoms.





 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download