Abstract
Liquid biopsy is a non-invasive method for tumor genotyping through detecting the circulating tumor DNA (ctDNA). Here, we describe the case of an esophageal squamous cell cancer patient in which a germline CDKN2A mutation was identified incidentally through liquid biopsy. The preoperative sample analysis revealed a total of five alterations in CDKN2A, TP53, FAT1, and KMT2C genes using next-generation sequencing data. The CDKN2A p.R87W was confirmed as a germline mutation, which is likely a pathogenic variant revealed through peripheral leukocyte DNA analysis. The patient underwent esophagectomy and sequential adjuvant chemoradiation therapy. After the surgery, the variant allele frequencies of somatic variants tended to decrease throughout the treatment. In addition to the detection of somatic variants, ctDNA testing can also provide information on the germline cancer susceptibility variants.
초록
액체생검은 순환종양 DNA (circulating tumor DNA, ctDNA)를 비침습적으로 분석하여 종양의 돌연변이를 검출하는 방법이다. 본 연구진은 식도편평세포암 환자의 액체생검 분석 중 부수적으로 CDKN2A 유전자의 생식세포 변이를 발견한 증례를 보고하고자 한다. 수술 전 채혈한 검체로 차세대염기서열분석을 시행하여 CDKN2A, TP53, FAT1 및 KMT2C 유전자에서 다섯 가지 변이를 검출하였다. CDKN2A p.R87W 변이는 말초혈액 백혈구 DNA 분석을 통해 생식세포 변이임이 확인되었다. 환자는 식도절제술 후 순차적인 보조 화학방사선치료를 시행 받았다. 수술 후 체성 변이들은 치료 경과에 따라 variant allele frequency (VAF)가 감소하는 양상을 보였다. ctDNA 검사는 체성 변이를 검출하는 목적 외에도, 암 감수성의 생식세포 변이를 발견함으로써 추가적인 정보를 제공할 수 있다.
Non-invasive tumor genotyping using circulating tumor DNA (ctDNA), or liquid biopsy, has been used increasingly in the clinical oncology field for cancer diagnosis, monitoring, and assessing the eligibility of the targeted therapy. Since a large part of the cell-free DNA (cfDNA) in plasma originates from the non-tumor cells, such as leukocytes, next-generation sequencing (NGS) using cfDNA for somatic variants of a solid tumor can help identify the germline variants at approximately 50% (heterozygous) or 100% (homozygous) variant allele frequency (VAF). Therefore, using liquid biopsy, the germline heterozygous pathogenic variants of cancer susceptibility genes can be identified incidentally with about 50% VAF, whereas somatic variants typically exhibit lower VAFs [1-4].
The cyclin-dependent kinase inhibitor 2A (CDKN2A) gene (OM-IM 600160) is a tumor suppressor gene and encodes two proteins, p16INK4a and p14ARF, both of which are involved in the regulation of cell cycle pathways [5]. The somatic oncogenic alterations in CDKN2A have been discovered in multiple tumor types. CDKN2A germline mutation carriers are mainly predisposed to malignant melanoma, pancreatic cancer, head and neck cancer, and lung cancer [6].
Here, we describe the case of a patient with esophageal squamous cell cancer in which a germline CDKN2A mutation was identified incidentally through liquid biopsy.
A 60-year-old male patient was diagnosed with upper esophageal squamous cell carcinoma through endoscopic biopsy after visiting an outside hospital with a one-month history of dyspepsia and nausea. The patient had a medical history of premature atrial contraction and smoking history of 10 pack-years. He also had a family history of maternal breast cancer and biliary cancer. The clinical stage based on the preoperative imaging analysis was confirmed as cT3N2. With the patient’s consent, he was enrolled in an observational study to predict the postoperative recurrence of eso-phageal cancer using liquid biopsy.
The preoperative blood collection followed by NGS analysis targeting 27 genes associated with esophageal cancer, including FGFR1, TP53, NOTCH1, FAT1, PIK3CA, FAT3, EP300, NFE2L2, ZNF750, KMT2C, RB1, FAT2, PTCH1, KDM6A, FBXW7, CDKN2A, PTEN, CREBBP, NOTCH2, AJUBA, KEAP1, KMT2D, EGFR, SMAD4, SMARCA4, NOTCH3, and NSD1, was performed. cfDNA was extracted from the plasma, and the prepared libraries were sequenced using a NovaSeq 6000 System (Illumina, San Diego, CA, USA). The initial liquid biopsy revealed five alterations in CDKN2A, TP53, FAT1, and KMT2C genes. The CDKN2A variant [NM_000077.4: c.259C>T, p.Arg87Trp (p.R87W)] was confirmed as a germline heterozygous mutation upon analyzing the peripheral leukocyte-extracted DNA. The remaining four variants of TP53 p.Y163X (NM_000546.5: c.489C>A, p.Tyr163Ter), FAT1 p.T1963I (NM_005245.3: c.5888C>T, p.Thr1963Ile), FAT1 p.C3780Y (NM_005245.3:c.11339G>A, p.Cys3780Tyr), and KMT2C p.I3596V (NM_170606.2:c.10786A>G, p.Ile3596Val) were confirmed as somatic mutations (Fig. 1). The CDKN2A p.R87W variant in this patient has previously been reported in multiple individuals with sporadic or familial melanoma [7-10]. Experimental studies have shown that the missense variant inhibits the binding of p16INK4a and CDK4 proteins and reduces the cell cycle and oxidative regulatory function of p16INK4a [10-12]. Also, the variant (rs749714198) was identified rarely in the gnomAD database (heterozygous in two of 232,208 chromosomes). Alternative missense substitution at this site (CDKN2A p.R87P) is likely represented as a pathogenic, loss-of-function variant [13-16]. Considering the functional evidence (PS3), population frequency (PM2), alternative substitution (PM5), and interpretations of ClinVar reports (PP5, https://www.ncbi.nlm.nih.gov/clinvar/, Variation ID: 406707), we classified the germline CDKN2A p.R87W variant as likely pathogenic, based on the guidelines of the 2015 American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) [17].
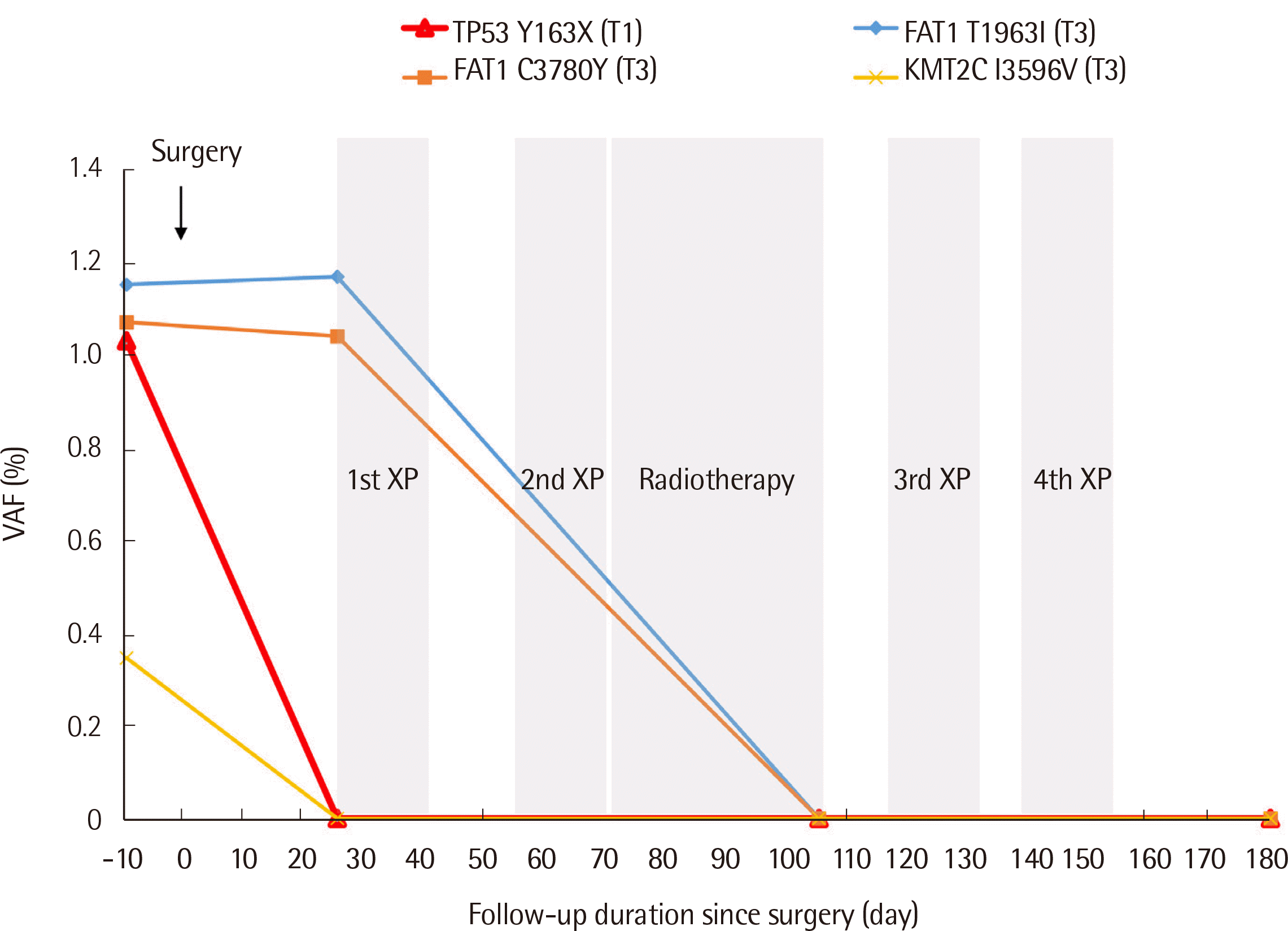
The patient underwent esophagectomy and total mediastinal lymph node dissection, while cervical esophagogastrostomy and pathologic reports revealed that he was at the final stage of upper esophageal cancer, poorly differentiated pT3N3, based on the 8th edition of the AJCC TNM cancer staging system [18]. The patient received sequential adjuvant chemoradiation therapy with cisplatin and capecitabine (XP). During nine months of follow-up, he exhibited no clinical or radiological signs of tumor recurrence or metastasis. Serial liquid biopsies were performed at 25, 105, and 180 days after surgery. A germline CDKN2A p.R87W variant with 50% of VAF was observed consistently even though there was no progression or recurrence of the postoperative disease. Other variants that were initially identified with a low VAF of 1.1–0.3%, including TP53 p.Y163X, FAT1 p.T1631I, FAT1 p.C3780Y, and KMT2C p.I3596V, exhibited a decreasing tendency throughout the treatment.
Germline CDKN2A mutations are common alterations found in 20-40% of cases with familial atypical multiple mole melanoma (FAMMM) syndrome [19]. Cancer risk elevation in mutation carriers is known in both familial and sporadic forms of various cancers, including pancreatic, head and neck, breast, and lung cancer [6, 19]. Our patient had no history of malignant melanoma or other cancers, but several studies have reported the relevance of CDKN2A mutation in esophageal cancer [19-21].
The NCCN guideline for genetic testing recommends pancreatic cancer screening for individuals with a known pathogenic/likely pathogenic variant of CDKN2A, beginning at the age 40 years. General melanoma risk management, such as annual skin examination and limiting UV exposure, is recommended for the individuals with germline pathogenic variants of CDKN2A [22]. In principle, genetic testing should be considered in high-risk individuals when the test result could impact the management plans for the tested family members. In our case, the patient has one son, and his living relatives include four brothers and three sisters, all without a history of cancer. The patient’s mother had breast cancer and biliary cancer, both of which were not classified to increase the risk for CDKN2A mutations, according to the NCCN guideline [22]. Although it is not indicated definitively in the guideline, the genetic screening for germline CDKN2A mutation, either through targeted sequencing or multi-gene testing, could be considered for the patient’s family members to assess their risk of developing cancer in future and establish the management plans accordingly.
There have been several reports on the identification of secondary germline mutations in cancer predisposition genes through ctDNA testing [1, 2, 4, 23]. Slavin et al. [1] recently reported an extensive data set on secondary germline cancer predisposition mutations in cancer patients who underwent commercial cfDNA testing. As a result, the germline hereditary cancer mutations were observed in 11 genes in 1.4% of patients. In the study, 10 CDKN2A germline mutation cases among 10,888 cases (0.09%) were identified, and there was no esophageal squamous cell cancer case with CDKN2A germline mutation [1]. Each institution should set a policy about the clinically significant germline findings regarding the consent, genetic counseling, and patient management [24, 25].
The other somatic variants in TP53, FAT, and KMT2C genes that were found preoperatively and during monitoring via liquid biopsy exhibited a decreasing trend throughout the treatment. This result in combination with the imaging and clinical findings, demonstrated the potential of cancer patient monitoring through ctDNA testing. Future studies should report the clinical implications of the feasibility of predicting disease progression or relapse in patients with esophageal cancer.
The limitation of our report is that we could not perform genetic testing in affected family members to confirm the causal role of the CDKN2A variant or penetrance data in the family. Nevertheless, this case is still relevant as it is the first in Korea to harbor the incidental germline cancer susceptibility variants identified using liquid biopsy. The detection of a pathogenic variant in a cancer susceptibility gene using liquid biopsy can provide information on the risk of cancer in patients and their families. The clinicians should be aware of the potential existence of incidental germline mutations and provide pre- and post -test genetic counseling to their patients.
REFERENCES
1. Slavin TP, Banks KC, Chudova D, Oxnard GR, Odegaard JI, Nagy RJ, et al. 2018; Identification of incidental germline mutations in patients with advanced solid tumors who underwent cell-free circulating tumor DNA sequencing. J Clin Oncol. JCO1800328. DOI: 10.1200/JCO.18.00328. PMID: 30339520. PMCID: PMC6286162.

2. Veyseh M, Ricker C, Espenschied C, Raymond V, D'Souza A, Barzi A. 2018; Secondary germline finding in liquid biopsy of a deceased patient; case report and review of the literature. Front Oncol. 8:259. DOI: 10.3389/fonc.2018.00259. PMID: 30050867. PMCID: PMC6052887.

3. Shukuya T, Patel S, Shane-Carson K, He K, Bertino EM, Shilo K, et al. 2018; Lung cancer patients with germline mutations detected by next-generation sequencing and/or liquid biopsy. J Thorac Oncol. 13:e17–9. DOI: 10.1016/j.jtho.2017.09.1962. PMID: 28989037. PMCID: PMC5910030.
4. Hu Y, Alden RS, Odegaard JI, Fairclough SR, Chen R, Heng J, et al. 2017; Discrimination of germline EGFR T790M mutations in plasma cell-free DNA allows study of prevalence across 31,414 cancer patients. Clin Cancer Res. 23:7351–9. DOI: 10.1158/1078-0432.CCR-17-1745. PMID: 28947568. PMCID: PMC5712272.
5. Sherr CJ. 2001; The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2:731–7. DOI: 10.1038/35096061. PMID: 11584300.

6. Chan SH, Chiang J, Ngeow J. 2021; CDKN2A germline alterations and the relevance of genotype-phenotype associations in cancer predisposition. Hered Cancer Clin Pract. 19:21. DOI: 10.1186/s13053-021-00178-x. PMID: 33766116. PMCID: PMC7992806.

7. Ruiz A, Puig S, Malvehy J, Lázaro C, Lynch M, Gimenez-Arnau AM, et al. 1999; CDKN2A mutations in Spanish cutaneous malignant melanoma families and patients with multiple melanomas and other neoplasia. J Med Genet. 36:490–3. PMID: 10874641. PMCID: PMC1734379.
8. Puig S, Potrony M, Cuellar F, Puig-Butille JA, Carrera C, Aguilera P, et al. 2016; Characterization of individuals at high risk of developing melanoma in Latin America: bases for genetic counseling in melanoma. Genet Med. 18:727–36. DOI: 10.1038/gim.2015.160. PMID: 26681309. PMCID: PMC4940430.

9. Helsing P, Nymoen DA, Ariansen S, Steine SJ, Maehle L, Aamdal S, et al. 2008; Population-based prevalence of CDKN2A and CDK4 mutations in patients with multiple primary melanomas. Genes Chromosomes Cancer. 47:175–84. DOI: 10.1002/gcc.20518. PMID: 18023021.
10. Kannengiesser C, Brookes S, del Arroyo AG, Pham D, Bombled J, Barrois M, et al. 2009; Functional, structural, and genetic evaluation of 20 CDKN2A germ line mutations identified in melanoma-prone families or patients. Hum Mutat. 30:564–74. DOI: 10.1002/humu.20845. PMID: 19260062.
11. Jenkins NC, Jung J, Liu T, Wilde M, Holmen SL, Grossman D. 2013; Familial melanoma-associated mutations in p16 uncouple its tumor-suppressor functions. J Invest Dermatol. 133:1043–51. DOI: 10.1038/jid.2012.401. PMID: 23190892. PMCID: PMC3594444.

12. Miller PJ, Duraisamy S, Newell JA, Chan PA, Tie MM, Rogers AE, et al. 2011; Classifying variants of CDKN2A using computational and laboratory studies. Hum Mutat. 32:900–11. DOI: 10.1002/humu.21504. PMID: 21462282.

13. Hussussian CJ, Struewing JP, Goldstein AM, Higgins PA, Ally DS, Shea-han MD, et al. 1994; Germline p16 mutations in familial melanoma. Nat Genet. 8:15–21. DOI: 10.1038/ng0994-15. PMID: 7987387.

14. Ranade K, Hussussian CJ, Sikorski RS, Varmus HE, Goldstein AM, Tucker MA, et al. 1995; Mutations associated with familial melanoma impair p16INK4 function. Nat Genet. 10:114–6. DOI: 10.1038/ng0595-114. PMID: 7647780.

15. Yu KK, Zanation AM, Moss JR, Yarbrough WG. 2002; Familial head and neck cancer: molecular analysis of a new clinical entity. Laryngoscope. 112:1587–93. DOI: 10.1097/00005537-200209000-00010. PMID: 12352668.

16. Reymond A, Brent R. 1995; p16 proteins from melanoma-prone families are deficient in binding to Cdk4. Oncogene. 11:1173–8. PMID: 7566978.
17. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. 2015; Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 17:405–24. DOI: 10.1038/gim.2015.30. PMID: 25741868. PMCID: PMC4544753.

18. Amin MB, Edge SB, editors. 2017. AJCC cancer staging manual. 8th ed. NewYork, NY: Springer International Publishing.
19. Middlebrooks CD, Stacey ML, Li Q, Snyder C, Shaw TG, Richardson-Nelson T, et al. 2019; Analysis of the CDKN2A gene in FAMMM syndrome families reveals early age of onset for additional syndromic cancers. Cancer Res. 79:2992–3000. DOI: 10.1158/0008-5472.CAN-18-1580. PMID: 30967399.
20. Lynch HT, Brand RE, Hogg D, Deters CA, Fusaro RM, Lynch JF, et al. 2002; Phenotypic variation in eight extended CDKN2A germline mutation familial atypical multiple mole melanoma-pancreatic carcinoma-prone families: the familial atypical mole melanoma-pancreatic carcinoma syndrome. Cancer. 94:84–96. DOI: 10.1002/cncr.10159. PMID: 11815963.
21. van der Wilk BJ, Noordman BJ, Atmodimedjo PN, Dinjens WNM, Laheij RJF, Wagner A, et al. 2020; Development of esophageal squamous cell cancer in patients with FAMMM syndrome: Two clinical reports. Eur J Med Genet. 63:103840. DOI: 10.1016/j.ejmg.2020.103840. PMID: 31923587.

22. Daly MB, Pal T, Berry MP, Buys SS, Dickson P, Domchek SM, et al. 2021; Genetic/familial high-risk assessment: breast, ovarian, and pancreatic, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 19:77–102. DOI: 10.6004/jnccn.2021.0001. PMID: 33406487.
23. Ratajska M, Koczkowska M, Żuk M, Gorczyński A, Kuźniacka A, Stukan M, et al. 2017; Detection of BRCA1/2 mutations in circulating tumor DNA from patients with ovarian cancer. Oncotarget. 8:101325–32. DOI: 10.18632/oncotarget.20722. PMID: 29254167. PMCID: PMC5731877.
24. Mandelker D, Donoghue M, Talukdar S, Bandlamudi C, inivasan P Sr, Vivek M, et al. 2019; Germline-focussed analysis of tumour-only sequencing: recommendations from the ESMO Precision Medicine Working Group. Ann Oncol. 30:1221–31. DOI: 10.1093/annonc/mdz136. PMID: 31050713. PMCID: PMC6683854.

25. Meric-Bernstam F, Brusco L, Daniels M, Wathoo C, Bailey AM, Strong L, et al. 2016; Incidental germline variants in 1000 advanced cancers on a prospective somatic genomic profiling protocol. Ann Oncol. 27:795–800. DOI: 10.1093/annonc/mdw018. PMID: 26787237. PMCID: PMC4843184.

26. Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. 2017; Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 19:4–23. DOI: 10.1016/j.jmoldx.2016.10.002. PMID: 27993330. PMCID: PMC5707196.
Fig. 1
Changes in the variant allele frequencies of somatic variants throughout the treatment. The somatic variants identified at the initial liquid biopsy exhibited a decreasing tendency throughout the treatment. The somatic variants were classified based on the 2017 Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists guidelines [26]. The variant allele frequency (VAF, %) was calculated as (read depth count of identified variant/total read depth count at the position)×100.
Abbreviations: T, Tier; VAF, variant allele frequency; XP, cisplatin and capecitabine chemotherapy.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download