Abstract
Candida auris is a multidrug-resistant fungal pathogen emerging worldwide that is closely related to the C. haemulonii species complex. The ASTA MicroIDSys (ASTA, Korea) is a new matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) system developed for species-level identification of microorganisms. However, prior to the current study, the reference database of ASTA MicroIDSys did not include C. auris. We expanded the database by adding 20 reference strains of C. auris and three closely related species belonging to C. haemulonii species complex. Further, we compared the performance of the ASTA system using an expanded database (coreDB v1.27.02) to that of the Biotyper system (Bruker Daltonics, USA) using 91 well-characterized Candida isolates from a Korean collection. In addition, we evaluated the ability of the ASTA system to differentiate between clade II and non-clade II isolates of C. auris. The results revealed that both ASTA and Biotyper systems accurately identified all 73 C. auris isolates. Of the 18 isolates of closely related species (nine C. pseudohaemulonii, seven C. haemulonii, and two C. haemulonii var. vulnera), the ASTA and Biotyper systems correctly identified 16 and 14 isolates, respectively, to the species level. Neither system misidentified any of the 91 isolates. Cluster analyses of the ASTA spectra distinctly discriminated clade II Korean C. auris isolates from the non-clade II isolates obtained from other countries. Our results show that the ASTA system with an expanded database is a reliable platform for the identification of C. auris and closely related species.
초록
Candida auris는 유전학적으로 Candida haemulonii complex 균종과 밀접히 연관되어 있으며, 전 세계적으로 새롭게 나타나고 있는 다제내성 진균이다. ASTA MicroIDSys (ASTA, Korea; 이하 “ASTA”라 한다)는 비교적 최근 개발된 MALDI-TOF 시스템으로서 각종 미생물을 동정하는 데 사용되고 있으나, 본 연구 전까지 데이터베이스에 C. auris가 포함되어 있지 않았다. 본 연구에서는 C. auris 및 C. haemulonii complex 3종에 속하는 20주의 표준 균주를 이용하여 ASTA 데이터베이스를 확장하고(coreDB v1.27.02), Bruker Biotyper (Bruker Daltonics, USA; 이하 “Biotyper”라 한다)와 함께 국내병원에서 분리된 Candida 91주에 대한 동정 성능을 비교하였다. 또한, ASTA가 C. auris clade II 균주와 non-clade II 균주를 구분할 수 있는 지 알아보았다. C. auris 73주는 ASTA와 Biotyper 두 시스템에 의해 모두 정확하게 동정되었고, 연관 균종 18주(Candida pseudohaemulonii 9주, Candida haemulonii 7주 및 Candida haemulonii var. vulnera 2주)는 ASTA와 Biotyper에 의해 각각 16주 및 14주가 정확히 동정되었다. 두 시스템에 의해 다른 균종으로 잘못 동정된 예는 한 주도 없었다. C. auris 균주들에 대한 ASTA의 스펙트럼을 이용하여 군집분석을 시행한 결과, 국내 분리 clade II C. auris 균주는 다른 나라에서 분리된 non-clade II 균주와 명확하게 구분되었다. 본 연구에서는 확장된 데이터베이스를 이용한 ASTA MALDI-TOF 시스템이 C. auris와 연관 균종을 동정하는 데 있어 신뢰할 만한 성능을 보여줌을 확인하였다.
A new matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS)-based ASTA MicroIDSys system (ASTA Inc, Suwon, Korea; hereafter, ASTA) has been developed and is being used for the identification of clinically important pathogens in microbiology laboratories [1-4]. The current ASTA MicroIDSys database (coreDB ver 1.27; hereafter, AMDB) contains 428 yeast species but does not include a reference spectrum database of Candida auris. C. auris was first reported in 2009 after isolation from ear cultures of a single Japanese patient and 15 Korean patients [5, 6]. Since then, C. auris has emerged as a multidrug-resistant healthcare-associated fungal pathogen globally [7-10]. C. auris can be grouped into four distinct clades, which are named in accordance with the locations wherein C. auris was initially isolated: South Asian (clade I), East Asian (clade II), African (clade III), and South American (clade IV) [9-11]. A recent Korean study found that C. auris isolates from Korean hospitals belong to clade II, which appear to have a propensity to affect the ear that is uncharacteristic of the other clades; other clades typically cause invasive infections and large-scale outbreaks [11, 12]. Therefore, in addition to accurate identification of C. auris, it may be essential to differentiate between clade II and non-clade II isolates of C. auris to ensure proper antifungal therapy and infection control measures. In this study, we expanded the AMDB using international reference strains of C. auris and closely related species; we also evaluated the effectiveness of the ASTA system after database expansion for identifying clinical isolates of C. auris and closely related species, compared to the Bruker Biotyper MS system (Bruker Daltonics, Billerica, MA, USA; hereafter, Biotyper). Further, we examined whether the ASTA system can differentiate clade II Korean isolates of C. auris from non-clade II strains from other countries.
The AMDB was expanded using 20 reference strains, including 15 strains from the U.S. Food and Drug Administration-Centers for Disease Control and Prevention (CDC) Antibiotic Resistance (AR) Isolate Bank (10 isolates from all four clades of C. auris [AR0381–AR0390], two C. haemulonii strains [AR0393 and AR0395], and three C. duobushaemulonii strains [AR0391, AR0392, and AR0394]), and five strains from the Korean Collection for Type Cultures (KCTC; two C. auris [KCTC 17809 and KCTC 17810], two C. pseudohaemulonii strains [KCTC 17806 and KCTC 17807], and one C. haemulonii [KCTC 17808] strains). All strains were inoculated onto potato dextrose agar plates and incubated at 20–25°C for 48 hours prior to spectra acquisition. The ASTA protocol was used for database expansion. Further, the on-plate formic acid (FA) extraction method was used for target preparation [2]. For each sample, colonies were prepared on 20 individual spots as technical replicates, and consequently 20 MALDI-TOF mass spectra were acquired. Each mass spectrum was obtained using 1,200 laser pulses. Peaks were introduced, ranging from 3,000 to 20,000 m/z. Further, the qualities of mass spectra were confirmed and each group of spectra was combined into a single mass spectrum for each group of replicates, called the reference spectrum. The reference spectrum is standardized data wherein m/z and intensities were averaged for frequently existing peaks in every replicate of the mass spectrum. Reference spectra were imported into the database for further analyses.
To compare the ASTA system after database expansion (coreDB v1.27.02) and the Biotyper system with library version 4.0, we tested a panel of molecularly identified Candida strains from a Korean collection. The Biotyper library version 4.0 has a database of only three strains of C. auris, two from Korea (KCTC 17809 and KCTC 17810) and one from Japan (DSM 21092T). The Korean collection consisted of 91 patient isolates including 73 C. auris, nine C. pseudohaemulonii, seven C. haemulonii, and two C. haemulonii var. vulnera isolates, which were submitted from 1996 to 2018 to the Chonnam National University Hospital, Gwangju, Korea, from 16 Korean hospitals [6, 12]. All isolates were identified via sequencing of the internal transcribed spacer (ITS) region and/or D1/D2 regions of the 26S ribosomal DNA of rRNA genes [6, 13]. Multilocus sequencing typing (MLST) of 73 isolates of C. auris was performed using a previously described method using four housekeeping genes (RPB1, RPB2, ITS, and D1/D2) [12]. For all 91 isolates, on-plate FA extraction was performed for identification using the ASTA system, whereas both on-plate FA extraction and in-tube FA plus acetonitrile (FA/ACN) extraction were performed for identification using the Biotyper system [2, 14]. The results from the two MALDI-TOF MS systems were compared to those of sequence-based identification and assigned to one of four categories: i) correct identification (identical to sequence-based identification with a cutoff score ≥140 [ASTA] or ≥1.7 [Biotyper]), ii) incomplete identification, iii) misidentification (either one incorrect species was identified or two or three incorrect species were proposed), or iv) no identification. Identification scores ≥1.7 of the Biotyper system were interpreted as acceptable identification at the species complex or species level, according to the findings of several previous reports [14-16], whereas cutoff scores ≥140 of the ASTA system were considered acceptable for species identification, according to the manufacturer’s recommendations [1, 2]. The ‘incomplete’ category was defined as a case in which two different species were proposed, and one was correct, or the isolate was correctly identified, but only to the C. haemulonii complex level (C. haemulonii, C. duobushaemulonii, and C. haemulonii var. vulnera). The ‘no identification’ category included cases with a score below the cutoff levels of 140 (ASTA) or 1.7 (Biotyper).
Table 1 shows the identification results and extraction methods for the two MALDI-TOF MS systems. Of the 91 isolates tested, 97.8% were correctly identified by the ASTA system to the species level after on-plate FA extraction and 95.6% were identified by the Biotyper system after in-tube FA/ACN extraction. The results indicated that the ASTA system has a comparable identification performance to the Biotyper system for isolates of C. auris and closely related species. The ASTA system correctly identified all 73 C. auris, nine C. pseudohaemulonii, and seven C. haemulonii isolates from the Korean collection to the species level after on-plate FA extraction. Two isolates of C. haemulonii var. vulnera, a strain that was not included in the AMDB, were incompletely identified as C. haemulonii. After in-tube FA/ACN extraction, the Biotyper system correctly identified all 73 isolates of C. auris; however, it correctly identified 77.8% (14/18) isolates of three closely related species. Two isolates of C. haemulonii var. vulnera and one isolate of C. haemulonii were not identified completely because two species (C. haemulonii or C. haemulonii var. vulnera) were proposed by the Biotyper system. Further, one isolate of C. pseudohaemulonii was not identified completely because two speci es (C. pseudohaemulonii or C. duobushaemulonii) were proposed by the Biotyper system. Even the additional use of the “10% rule”, which excluded the result scoring >10% below the top-scoring match [17], did not resolve any incomplete identification results.
In-tube FA/ACN extraction generates best-quality mass spectra, but on-plate FA extraction is a simpler method that may be generally useful for clinical purposes [18]. Only 79.1% (72/91) were correctly identified when we attempted to identify all isolates using the Biotyper system after on-plate FA extraction. Of the 19 isolates that were not correctly identified after on-plate FA extraction, only two isolates of C. pseudohaemulonii were correctly identified with the use of the “10% rule.” This indicated that the Biotyper system tends to have better results following the in-tube FA/ACN method for C. auris and closely related species, in agreement with a previous report [19]. There were no misidentification results when the Biotyper system was used after on-plate FA extraction, suggesting that repeat Biotyper testing with additive in-tube FA/ACN extraction for isolates with “incomplete or no identification” results after initial on-plate FA extraction may provide a cost-effective and reliable method to identify these isolates. The reasons for the low rate of correct C. auris identification by the Biotyper after on-plate FA extraction are poorly understood, but may be due to the characteristics of this pathogen, which renders solubilization of proteins difficult by simple extraction on the plate, or due to insufficient database entries to enable spectral matches, as the C. auris database is designed for isolates after in-tube FA/ACN extraction [12].
The recent emergence of C. auris has drawn attention to closely related species, including the C. haemulonii complex and C. pseudohaemulonii [9]. Despite these closely related species being considered rare human pathogens, the increasing rate of incidence and antifungal resistance associated with these species present a cause for concern worldwide [13]. The present study demonstrates that although excellent results were obtained with the use of ASTA and Biotyper systems for the identification of C. auris, the systems are somewhat limited in their abilities to differentiate between C. haemulonii and C. haemulonii var. vulnera. However, neither system returned any misidentification results, enabling these closely related species to be distinguished by sequence-based identification.
In a previous study, we showed that MLST analyses can differentiate C. auris reference isolates of the four clades (Clade I–IV) into four clusters (ST clusters 1–4); we also found that all Korean isolates of C. auris have the same MLST type as ST cluster 2 (clade II) [12]. In the present study, 72 out of 73 isolates tested also showed identical MLST type, ST cluster 2 (profile of four alleles ITS-RPB1-RPB2-D1/D2, a - a - a - a), whereas one isolate that was discovered in 2018 was classified as ST cluster 1 (profile of four alleles ITS-RPB1-RPB2-D1/D2, b - b - a - b). Fig. 1 depicts the result of cluster analyses of the MS profiles, based on peak similarity, generated from the 17 C. auris (K1 to K17) isolates from Korean hospitals and the 20 international reference strains used to expand the AMDB. A dendrogram shows the clear discrimination of C. auris from the other three related species. Notably, 16 Korean isolates of C. auris (K1 to K16, ST cluster 2) and one reference isolate of clade II (C. auris AR0381) were clustered with C. auris clade II. One Korean isolate of C. auris (K17), which belonged to ST cluster 1, was clustered with the non-clade II group isolates of C. auris consisting of nine CDC isolates of C. auris from three different geographic clades (clades I, III, and IV). Therefore, to the best of our knowledge, this is the first study to prove the potential of a MALDI-TOF MS (ASTA) system in differentiating clade II from non-clade II C. auris isolates. Given that multiple introductions and subsequent transmission of C. auris in the US have been reported on the basis of genotypes [20], further molecular epidemiologic study is needed for the one ST cluster 1 isolate of C. auris discovered from a Korean hospital in 2018.
In summary, here, we constructed an expanded clinical database for the ASTA system (coreDB v1.27.02) using 20 reference strains of C. auris and three closely related species that facilitated the successful identification of those species. Although coreDB v1.27.02 of the ASTA system cannot differentiate between C. haemulonii and C. haemulonii var. vulnera, species-level identification will be improved in the near future through further database expansion using internationally available strains of C. haemulonii var. vulnera.
Acknowledgments
ASTA supplied the MALDI plate and matrix solution free of charge. We thank the U.S. CDC for kindly providing a panel of C. auris and closely related isolates.
REFERENCES
1. Lee Y, Sung JY, Kim H, Yong D, Lee K. 2017; Comparison of a new matrix-assisted laser desorption/ionization time-of-flight mass spectrometry platform, ASTA MicroIDSys, with Bruker Biotyper for species identification. Ann Lab Med. 37:531–5. DOI: 10.3343/alm.2017.37.6.531. PMID: 28840993. PMCID: PMC5587828.

2. Lee H, Park JH, Oh J, Cho S, Koo J, Park IC, et al. 2019; Evaluation of a new matrix-assisted laser desorption/ionization time-of-flight mass spectrometry system for the identification of yeast isolation. J Clin Lab Anal. 33:e22685. DOI: 10.1002/jcla.22685. PMID: 30298531. PMCID: PMC6818552.

3. Kim D, Ji S, Kim JR, Kim M, Byun JH, Yum JH, et al. 2020; Performance evaluation of a new matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, ASTA MicroIDSys system, in bacterial identification against clinical isolates of anaerobic bacteria. Anaerobe. 61:102131. DOI: 10.1016/j.anaerobe.2019.102131. PMID: 31778809.

4. Jung J, Kim SY, Park YJ, Lee J, Suk HS, Ha SI, et al. 2020; Comparison of the ASTA MicroIDSys and VITEK MS matrix-assisted laser desorption/ionization time-of-flight mass spectrometry systems for identification of clinical bacteria and yeasts. J Infect Chemother. 26:1328–33. DOI: 10.1016/j.jiac.2020.08.004. PMID: 32855038.

5. Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. 2009; Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 53:41–4. DOI: 10.1111/j.1348-0421.2008.00083.x. PMID: 19161556.
6. Kim MN, Shin JH, Sung H, Lee K, Kim EC, Ryoo N, et al. 2009; Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin Infect Dis. 48:e57–61. DOI: 10.1086/597108. PMID: 19193113.
7. Lee WG, Shin JH, Uh Y, Kang MG, Kim SH, Park KH, et al. 2011; First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol. 49:3139–42. DOI: 10.1128/JCM.00319-11. PMID: 21715586. PMCID: PMC3165631.
8. Abastabar M, Haghani I, Ahangarkani F, Rezai MS, Taghizadeh Armaki M, Roodgari S, et al. 2019; Candida auris otomycosis in Iran and review of recent literature. Mycoses. 62:101–5. DOI: 10.1111/myc.12886. PMID: 30585653.
9. Gade L, Muñoz JF, Sheth M, Wagner D, Berkow EL, Forsberg K, et al. 2020; Understanding the emergence of multidrug-resistant Candida: using whole-genome sequencing to describe the population structure of Candida haemulonii species complex. Front Genet. 11:554. DOI: 10.3389/fgene.2020.00554. PMID: 32587603. PMCID: PMC7298116.

10. Chow NA, de Groot T, Badali H, Abastabar M, Chiller TM, Meis JF. 2019; Potential fifth clade of Candida auris, Iran, 2018. Emerg Infect Dis. 25:1780–1. DOI: 10.3201/eid2509.190686. PMID: 31310230. PMCID: PMC6711235.
11. Welsh RM, Sexton DJ, Forsberg K, Vallabhaneni S, Litvintseva A. 2019; Insights into the unique nature of the East Asian clade of the emerging pathogenic yeast Candida auris. J Clin Microbiol. 57:e00007–19. DOI: 10.1128/JCM.00007-19. PMID: 30760535. PMCID: PMC6440783.

12. Kwon YJ, Shin JH, Byun SA, Choi MJ, Won EJ, Lee D, et al. 2019; Candida auris clinical isolates from South Korea: identification, antifungal susceptibility, and genotyping. J Clin Microbiol. 57:e01624–18. DOI: 10.1128/JCM.01624-18. PMID: 30728190. PMCID: PMC6440790.

13. Lima SL, Francisco EC, de Almeida Júnior JN, Santos DWCL, Carlesse F, Queiroz-Telles F, et al. 2020; Increasing prevalence of multidrug-resistant Candida haemulonii species complex among all yeast cultures collected by a reference laboratory over the past 11 years. J Fungi (Basel). 6:110. DOI: 10.3390/jof6030110. PMID: 32679832. PMCID: PMC7558365.
14. Lee HS, Shin JH, Choi MJ, Won EJ, Kee SJ, Kim SH, et al. 2017; Comparison of the Bruker Biotyper and VITEK MS matrix-assisted laser desorption/ionization time-of-flight mass spectrometry systems using a formic acid extraction method to identify common and uncommon yeast isolates. Ann Lab Med. 37:223–30. DOI: 10.3343/alm.2017.37.3.223. PMID: 28224768. PMCID: PMC5339094.

15. Pence MA, McElvania TeKippe E, Wallace MA, Burnham CA. 2014; Comparison and optimization of two MALDI-TOF MS platforms for the identification of medically relevant yeast species. Eur J Clin Microbiol Infect Dis. 33:1703–12. DOI: 10.1007/s10096-014-2115-x. PMID: 24800928.

16. Cassagne C, Cella AL, Suchon P, Normand AC, Ranque S, Piarroux R. 2013; Evaluation of four pretreatment procedures for MALDI-TOF MS yeast identification in the routine clinical laboratory. Med Mycol. 51:371–7. DOI: 10.3109/13693786.2012.720720. PMID: 22978312.

17. Khot PD, Couturier MR, Wilson A, Croft A, Fisher MA. 2012; Optimization of matrix-assisted laser desorption ionization-time of flight mass spectrometry analysis for bacterial identification. J Clin Microbiol. 50:3845–52. DOI: 10.1128/JCM.00626-12. PMID: 22993178. PMCID: PMC3502975.

18. Fraser M, Brown Z, Houldsworth M, Borman AM, Johnson EM. 2016; Rapid identification of 6328 isolates of pathogenic yeasts using MALDI-ToF MS and a simplified, rapid extraction procedure that is compatible with the Bruker Biotyper platform and database. Med Mycol. 54:80–8. DOI: 10.1093/mmy/myv085. PMID: 26591008.

19. Mizusawa M, Miller H, Green R, Lee R, Durante M, Perkins R, et al. 2017; Can multidrug-resistant Candida auris be reliably identified in clinical microbiology laboratories? J Clin Microbiol. 55:638–40. DOI: 10.1128/JCM.02202-16. PMID: 27881617. PMCID: PMC5277535.
20. Chow NA, Gade L, Tsay SV, Forsberg K, Greenko JA, Southwick KL, et al. 2018; Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: a molecular epidemiological survey. Lancet Infect Dis. 18:1377–84. DOI: 10.1016/S1473-3099(18)30597-8. PMID: 30293877. PMCID: PMC6556114.
Fig. 1
Heat map (A) and dendrogram (B) of mass spectrometry (MS) profiles generated by the ASTA system for 17 representative C. auris isolates (K1 to K17) from Korean hospitals and 20 internationally collected reference strains of C. auris and closely related species used for database expansion of the ASTA system (AR0381–AR0395, KCTC 17806–17810). The isolation year, source, and MLST type of the Korean isolates (K1 to K17) are shown following the species name (C. auris) of each isolate. (A) Columns show each strain of the species, rows indicate clustered m/z, and heat represents the degree of intensity. (B) The dendrogram is constructed by hierarchical clustering and is labeled with strain names. Strains were clustered by species, and strains of C. auris were clustered into clade II and non-clade II groups.
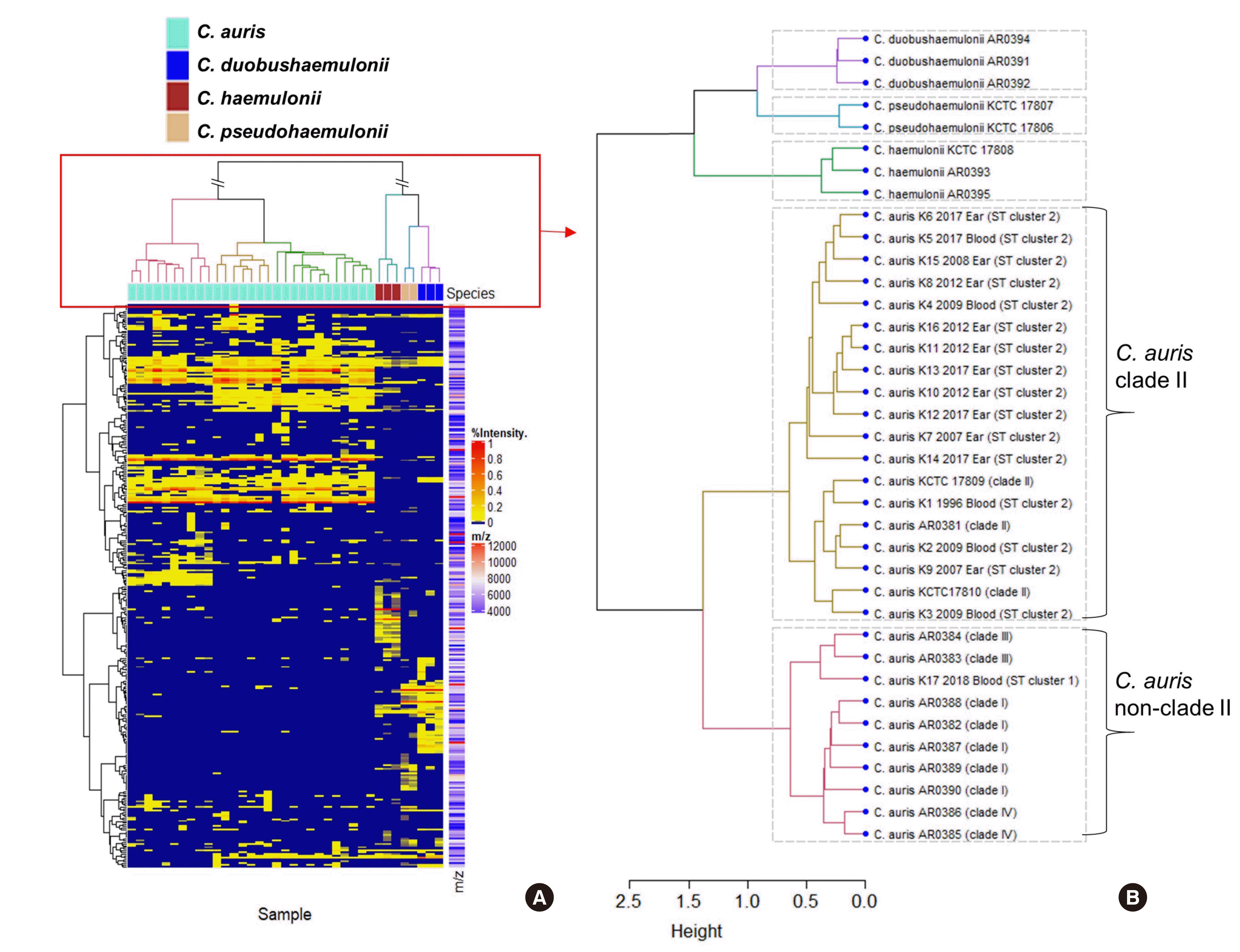
Table 1
Identification results of 91 isolates of Candida auris and three closely related species using two MALDI-TOF MS systems
| System (database) | Extraction method* | Species (N of isolates tested) | No. (%) of isolates† | |||
|---|---|---|---|---|---|---|
|
|
||||||
| Correct ID | Incomplete ID | No ID | Mis-ID | |||
| ASTA (CoreDB v1.27.02) | On-plate FA | C. auris (73) | 73 (100) | 0 (0) | 0 (0) | 0 (0) |
| C. pseudohaemulonii (9) | 9 (100) | 0 (0) | 0 (0) | 0 (0) | ||
| C. haemulonii (7) | 7 (100) | 0 (0) | 0 (0) | 0 (0) | ||
| C. haemulonii var. vulnera (2) | 0 (0) | 2 (100) | 0 (0) | 0 (0) | ||
| Total (91) | 89 (97.8) | 2 (2.2) | 0 (0) | 0 (0) | ||
| Biotyper (library version 4.0) | In-tube FA/ACN | C. auris (73) | 73 (100) | 0 (0) | 0 (0) | 0 (0) |
| C. pseudohaemulonii (9) | 8 (88.9) | 1 (11.1) | 0 (0) | 0 (0) | ||
| C. haemulonii (7) | 6 (85.7) | 1 (14.3) | 0 (0) | 0 (0) | ||
| C. haemulonii var. vulnera (2) | 0 (0) | 2 (100) | 0 (0) | 0 (0) | ||
| Total (91) | 87 (95.6) | 4 (4.4) | 0 (0) | 0 (0) | ||
| Biotyper (library version 4.0) | On-plate FA | C. auris (73) | 61 (83.6) | 0 (0) | 12 (16.4) | 0 (0) |
| C. pseudohaemulonii (9) | 7 (77.8) | 2 (22.2) | 0 (0) | 0 (0) | ||
| C. haemulonii (7) | 4 (57.1) | 2 (28.6) | 1 (14.3) | 0 (0) | ||
| C. haemulonii var. vulnera (2) | 0 (0) | 2 (100) | 0 (0) | 0 (0) | ||
| Total (91) | 72 (79.1) | 6 (6.6) | 13 (14.3) | 0 (0) | ||
*The on-plate formic acid (FA) extraction was performed for identification using the ASTA and Biotyper systems, whereas in-tube formic acid plus acetonitrile (FA/ACN) extraction was performed for identification using the Biotyper system; †The category includes correct ID (identical to sequence-based identification) to species level with cutoff scores ≥140 by the ASTA system and ≥1.7 by the Biotyper system. The “incomplete ID” category includes i) a “low level of discrimination” (two species were proposed, and one was correct), ii) correct ID to the C. haemulonii species complex level.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download