INTRODUCTION
METHODS
Cell culture
Cell viability test
Alizarin Red S staining
Quantitative PCR and quantitative real time-PCR
Table 1
Table 2
Immunoblotting
Statistical analysis
RESULTS
NAMPT expression was up-regulated during cell differentiation of MDPC-23 odontoblastic-like cells
 | Fig. 1The expression of NAMPT during odontoblastic differentiation of MDPC-23 cells.(A) Mineralized nodule formation in MDPC-23 cells. MDPC-23 cells were cultured with odontoblastic induction medium for 10 days, and mineralization was evaluated by alizarin Red S staining (upper panel). The mineralization was quantified by colorimetric spectrophotometry (lower panel). (B) mRNAs were prepared on day 0, 4, 7, and 10 after induction. qPCR was performed to detect the expression levels of ALP, Col-1, DMP-1, DSPP, BSP, and NAMPT. Quantitative data for mRNA expressions (B, upper panel) were analyzed by using ImageJ software after GAPDH normalization (B, lower panel). Each data point represents the mean ± SEM of three experiments. NAMPT, nicotinamide phosphoribosyltransferase; MDPC-23, mouse dental papilla cell-23; BSP, bone sialoprotein; DMP-1, dentin matrix protein-1; DSPP, dentin sialophosphoprotein; ALP, alkaline phosphatase; Col-1, collagen type-1; GAPDH, glyceraldehyde 3-phosphate dehydrogenase. *p < 0.05, **p < 0.01, ***p < 0.001.
|
NAMPT increased mineralization in MDPC-23 odontoblasts
 | Fig. 2The effects of Visfatin on mineralization in MDPC-23 cells.(A) MDPC-23 cell viability after Visfatin treatment as determined by MTT assays. (B) Alizarin Red S stain (upper panel) showing mineralized nodule formation in MDPC-23 cells, cultured in differentiation media and treated with Visfatin for up to 10 days. The mineralization was quantified using colorimetric spectrophotometry (lower panel). (C) Treatment with Visfatin at 50 ng/ml according to the defined conditions. The expression level of odontoblast biomarkers were assayed by Western blotting according to standard methods. Quantitative data for protein expressions (C, upper panel) were analyzed by using ImageJ software after β-actin normalization (C, lower panel). Each data point represents the mean ± SEM of three experiments. MDPC-23, mouse dental papilla cell-23; DSPP, dentin sialophosphoprotein; DMP-1, dentin matrix protein-1; NAMPT, nicotinamide phosphoribosyltransferase; BSP, bone sialoprotein. *p < 0.05, **p < 0.01, ***p < 0.001.
|
NAMPT inhibitor FK866 decreased odontoblast formation in MDPC-23 odontoblasts
 | Fig. 3The effects of the specific NAMPT inhibitor, FK866, on mineralization in MDPC-23 cells.(A) MDPC-23 cell viability after FK866 treatment as determined by MTT assays. (B) Alizarin Red S stain (left panel) showing mineralized nodule formation in MDPC-23 cells, cultured in differentiation media and treated with FK866 for up to 10 days. The mineralization was quantified using colorimetric spectrophotometry (right panel). Each data point represents the mean ± SEM of three experiments. NAMPT, nicotinamide phosphoribosyltransferase; MDPC-23, mouse dental papilla cell-23. *p < 0.05, **p < 0.01, ***p < 0.001.
|
 | Fig. 4The role of the specific NAMPT inhibitor, FK866, on differentiation in MDPC-23.qRT-PCR was performed to detect the expression levels of BSP (A), ALP (B), Col-1 (C), DMP-1 (D), DSPP (E), and NAMPT (F). GAPDH served as the experimental control. All data are based on three independent experiments. (G) Treatment with FK866 at 1 nM according to the defined conditions. Western blotting was used to assess the expression level of odontoblast biomarkers. Quantitative data for protein expressions (G, left panel) were analyzed by using ImageJ software after β-actin normalization (G, right panel). Each data point represents the mean ± SEM of three experiments. MDPC-23, mouse dental papilla cell-23; BSP, bone sialoprotein; ALP, alkaline phosphatase; Col-1, collagen type-1; DMP-1, dentin matrix protein-1; DSPP, dentin sialophosphoprotein; NAMPT, nicotinamide phosphoribosyltransferase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase. *p < 0.05, **p < 0.01, ***p < 0.001.
|
NAMPT regulates the Runx signal during MDPC-23 odontoblast differentiation
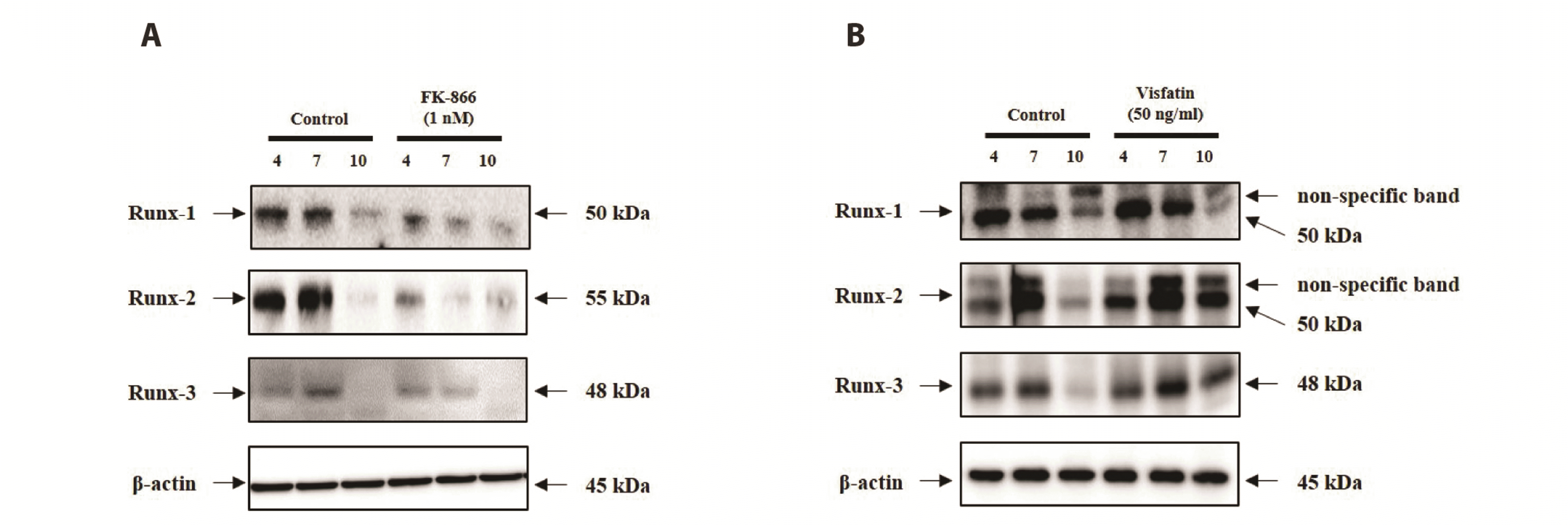 | Fig. 5Analysis of the NAMPT synthesis pathway in MDPC-23 odontoblast differentiation.Western blotting was performed to detect the protein expression levels of Runx-1, Runx-2, and Runx-3 in MDPC-23 cells treated with FK866 (A) or Visfatin (B). β-Actin served as the experimental control. The expression level of Runx family proteins were assayed using Western blotting. NAMPT, nicotinamide phosphoribosyltransferase; MDPC-23, mouse dental papilla cell-23.
|




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download