Dear Editor,
The main resistance mechanism of carbapenem-resistant
Enterobacteriaceae (CRE) is through acquired carbapenemases [
1]. Carbapenemase-producing
Enterobacteriaceae (CPE) harboring plasmid-encoded resistance genes easily spread the genes to different species [
2]. A screening test for CPE should be performed for infection control and prevention [
3]. The Korea Disease Control and Prevention Agency conducted a surveillance beginning in June 2017 [
4]. A European study suggests that within-hospital transmission and inter-hospital spread of CPE is more frequent within countries rather than between countries [
5]. Therefore, both hospital and national surveillance are important to help prevent the CPE spread .
We retrospectively analyzed active surveillance data for CRE at Hanyang University Seoul Hospital, Seoul, Korea, from July 2017 to December 2020. The candidates for CRE surveillance were patients who were transferred from long-term care facilities in the previous three months, those admitted to the intensive care unit, and those who had positive results for CRE isolates in the previous six months. Stool or rectal swab samples were inoculated onto chromID CARBA medium (bioMérieux, Marcy l’Etoile, France) and incubated at 35°C for 24 hours. Bacterial species were identified using matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) with a MALDI Biotyper (Bruker Daltonics, Bremen, Germany). Antimicrobial susceptibility testing was performed using the MicroScan WalkAway system (Beckman Coulter, Brea, CA, USA). Screening for carbapenemase-producing organisms was carried out according to the Clinical Laboratory Standards Institute guidelines using the modified Hodge test (MHT) during 2017–2018 and the modified carbapenem inactivation method (mCIM) during 2019–2020 [
6]. The carbapenemase inhibition test (CIT) using phenylboronic acid and EDTA was performed in conjunction with the MHT or mCIM [
7]. Carbapenemase genes were detected using the Xpert Carba-R assay (Cepheid, Sunnyvale, CA, USA). The study was approved by the Hanyang University Hospital Institute Review Board, Seoul, Korea (IRB number 202105037).
During the four years of the study period, 10,174 surveillance tests were conducted. Among the patients tested, 229 (2.3%) were new cases that carried CRE and 182 (1.8%) were positive for CPE. The CRE trends were analyzed quarterly from 2017 to 2020. Trends in the number of CRE screening tests that identified CRE or CPE cases are shown in
Fig. 1. The average number of CRE screening tests per three months was 727 (range: 580–893), including an average of 16 CRE (range: 9–30) and 10 CPE cases (range: 3–19). The CRE-positive rate was 2.3% (range: 1.1%–4.3%). Interestingly, the positive rates were highest in the third quarters of 2018 and 2020. Among the 229 total CRE isolates,
Klebsiella pneumoniae (55.0%) was predominant, followed by
Escherichia coli (28.4%), other
Klebsiella spp. (8.3%),
Citrobacter spp. (4.4%), and
Enterobacter spp. (3.5%).
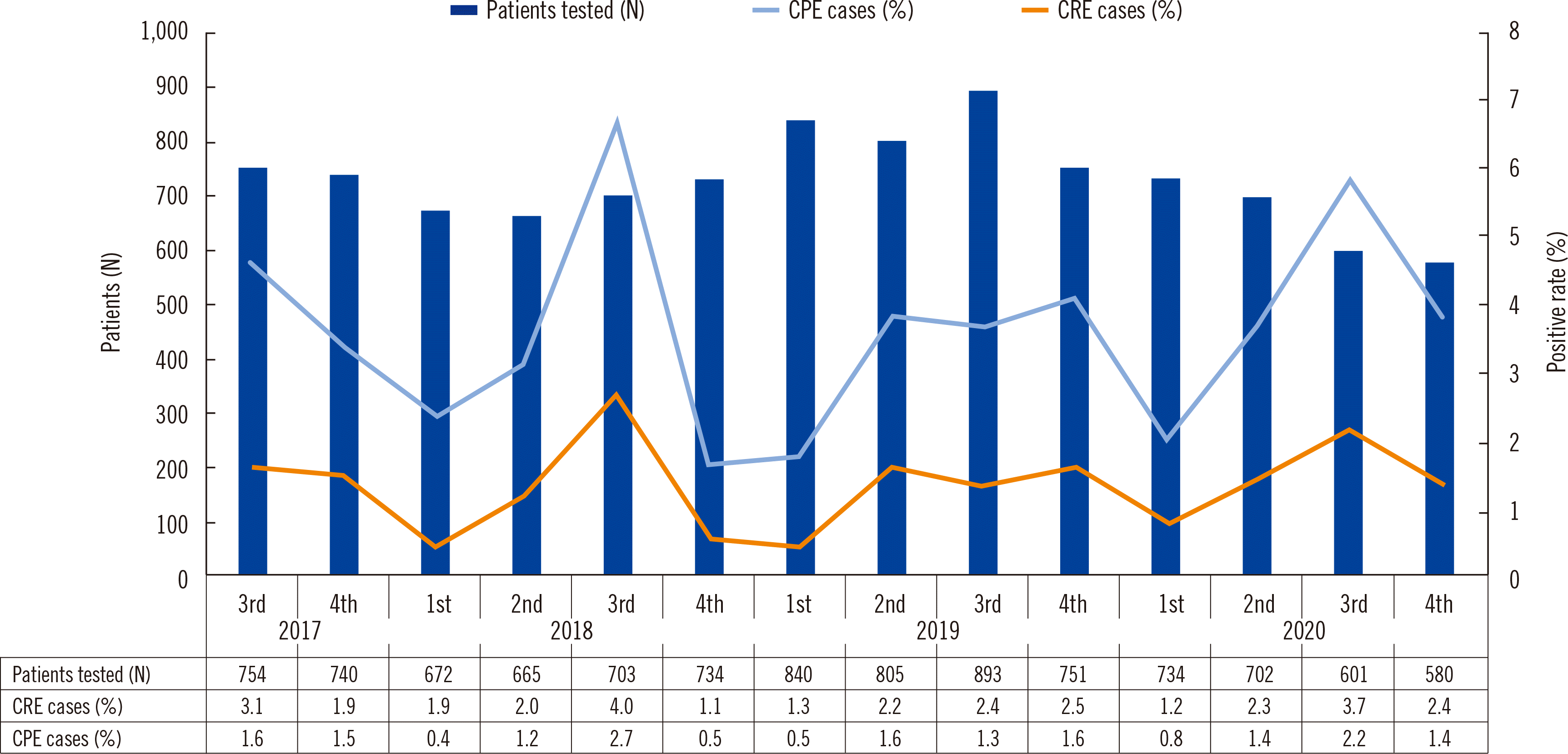 | Fig. 1Quarterly trends in the number of carbapenem-resistant Enterobacteriaceae (CRE) screening tests administered and the rates of positive CRE and carbapenemase-producing Enterobacteriaceae (CPE) cases from July 2017 to December 2020. 
|
Carbapenemase screening using MHT or mCIM was performed on 229 CRE isolates (
Table 1), of which, 147 isolates (64.2%) were positive and 82 (35.8%) were negative. The CIT results revealed that 151 isolates (65.9%) were positive. Carbapenemase-specific PCR was carried out for 178 CRE isolates, with carbapenemase genes being detected in 75.8% (135/178) of the isolates. Among the 135 isolates, 93 harbored
blaKPC (68.9%), which was the most prevalent gene, followed by 34 isolates with
blaNDM (25.2%) and two isolates with
blaOXA-48 (1.5%). These data are very similar to the 2018–2019 Korean national data of 70.0% for
blaKPC and 24.0% for
blaNDM [
4]. However, global surveillance has revealed that
blaKPC is the most common gene at 53.18%, followed by the
blaOXA-48-like gene at 20.09% and
blaNDM at 19.42% [
8]. Among the 72 carbapenemase-producing
K. pneumoniae isolates, 63 (87.5%) harbored
blaKPC while six (8.3%) carried
blaNDM. Meanwhile, among the 42 E. coli isolates, 20 (47.6%) carried
blaKPC and 18 (42.9%) carried
blaNDM.
Table 1
Carbapenemase screening and PCR testing of carbapenemase genes for identified CRE isolates
|
2017 |
2018 |
2019 |
2020 |
Total (%) |
|
MHT or mCIM* (N = 229) |
|
Positive |
24 |
35 |
47 |
41 |
147 (64.2) |
|
Negative |
13 |
27 |
22 |
20 |
82 (35.8) |
|
CIT (N = 229) |
|
Positive |
27 |
35 |
50 |
39 |
151 (65.9) |
|
Negative |
10 |
27 |
19 |
22 |
78 (34.1) |
|
Carbapenemase gene (N = 178) |
|
PCR Positive |
23 |
34 |
41 |
37 |
135 (75.8) |
|
blaKPC
|
14 |
26 |
27 |
26 |
93 (68.9) |
|
blaNDM
|
8 |
5 |
12 |
9 |
34 (25.2) |
|
blaIMP-1
|
0 |
0 |
1 |
1 |
2 (1.5) |
|
blaVIM
|
0 |
1 |
0 |
0 |
1 (0.7) |
|
blaOXA-48
|
0 |
1 |
0 |
0 |
1 (0.7) |
|
Other†
|
1 |
1 |
1 |
1 |
4 (3.0) |
|
PCR Negative |
11 |
14 |
10 |
8 |
43 (24.2) |
|
CP-K. pneumoniae (N = 72) |
|
blaKPC
|
10 |
19 |
17 |
17 |
63 (87.5) |
|
blaNDM
|
3 |
0 |
2 |
1 |
6 (8.3) |
|
blaIMP-1
|
0 |
1 |
0 |
0 |
1 (1.4) |
|
CP-E. coli (N = 42) |
|
|
|
|
|
|
blaKPC
|
1 |
5 |
7 |
7 |
20 (47.6) |
|
blaNDM
|
5 |
3 |
4 |
6 |
18 (42.9) |
|
blaOXA-48
|
0 |
0 |
1 |
1 |
2 (4.8) |

This study investigated the trends for CRE and CPE cases at a single hospital for the past four years. Our results were similar to the 1.4% rate reported in another study conducted in 2017 [
9]. CRE cases in this study may have exhibited seasonal and temperature variations, consistent with those shown in the study by Kim,
et al. [
10]. The current study has some limitations. First, the medical records of the patients were analyzed retrospectively. Second, we used only a chromogenic agar and a commercially available kit for detection of CPE. Therefore, CPE genes other than
blaKPC,
blaNDM,
blaVIM,
blaIMP, and
blaOXA-48 could not be detected.
In conclusion, the prevalence of CRE and CPE at the study hospital were 2.3% and 1.8%, respectively. The blaKPC gene was detected in 68.9% of the CPE isolates. Continuous monitoring for CPE is necessary to prevent the spread of CPE.
Go to :





 PDF
PDF Citation
Citation Print
Print




 XML Download
XML Download