Dear Editor,
In Korea, more than 130,000 cases of coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2), were recorded from January 1, 2020 to May 14, 2021, with approximately 600 cases diagnosed per day following published guidelines for the laboratory diagnosis of COVID-19 in Korea [1]. The number of SARS-CoV-2 variants has been increasing, posing a significant threat in terms of increased infectivity and clinical severity accompanied by reduced immunity upon vaccination [2-4].
Most SARS-CoV-2 variants arise due to changes in the N-terminal domain (NTD) and receptor-binding domain (RBD) [4, 5]. Among NTD variants, the spike deletion H69/V70 (del69-70), which exerts a marginal effect on 50% neutralization values in plaque reduction neutralization tests of post-vaccination serum samples, is present in the B.1.1.7 variant that was initially dominant in the United Kingdom (UK) before spreading worldwide [6]. N501Y is the most prominent RBD variant, which improves SARS-CoV-2 binding affinity, and is present in the UK (B.1.1.7), the Republic of South Africa (RSA) (B.1.351), and Brazil/Japan (P.1.) variants [4, 6]. In addition, K417N, which can improve the receptor binding of SARS-CoV-2, is present in the RSA variant B.1.351, and E484K, associated with immune escape from monoclonal antibodies, has been detected in both the RSA and Brazilian variants [4, 7, 8].
PowerChek SARS-CoV-2 S-gene Mutation Detection Kit (Kogene Biotech, Seoul, Korea) based on multiplex real-time reverse transcription (rRT)-PCR was developed for detecting COVID-19 variants, especially the UK (B.1.1.7), RSA (B.1.351), Brazil/Japan (P.1), and UK (B.1.525)/Brazil (P.2) variants. Using this kit, we evaluated the incidence of SARS-CoV-2 variants in the Ulsan area of Korea; to our knowledge, this is the first study to evaluate the incidence of SARS-CoV-2 variants in a specific area of Korea. This study was approved by the Institutional Review Board of Ulsan University Hospital, Ulsan, Korea (approval number: 2021-05-025).
Data from 36 SARS-CoV-2–positive respiratory samples, tested using PowerChek SARS-CoV-2 Real-time PCR Kit (Kogene Biotech) at our institution between April 1 and April 30, 2021, were assessed retrospectively. RNA samples were subjected to SARS-CoV-2 variant screening using PowerChek SARS-CoV-2 S-gene Mutation Detection Kit following the manufacturer’s instructions. Three mutation sites (N501Y, E484K, and K417N) were screened, and the results were classified as (1) UK (B.1.1.7) variant if only N501Y was detected, (2) Brazil/Japan (P.1) variant if both N501Y and E484K were detected, (3) RSA (B1.351) variant if all three variants were detected, and (4) UK (B1.525)/Brazil (P.2) variant if only E484K was detected. Direct sequencing of both the NTD (for the 69/70 deletion) and RBD was subsequently performed to confirm the detected variants, using primers designed in-house (for del69-70: forward 5´-CAC ACG TGG TGT TTA CCC T-3´, reverse 5´-GTT AGA CTT CTC AGT GGA AGC A-3´; for RBD: forward 5´-GAG GTG ATG AAG TCA GAC AAA TCG-3´, reverse 5´-CTC TGT ATG GTT GGT AAC CAA CA-3´). Examples of rRT-PCR and direct sequencing results of SARS-CoV-2 variants are presented in Fig. 1, and the results of all 36 samples are summarized in Table 1.
Using rRT-PCR, N501Y, E484K, and K417N were detected in 34 (94.4%), 1 (2.8%), and 0 (0.0%) samples, respectively, and the incidence rates of the B.1.1.7 (UK) and P.1 (Brazil/Japan) variants were calculated as 33/36 (91.7%) and 1/36 (2.8%), respectively. Using direct sequencing, del69-70, N501Y (A→T), E484K (G→A), and K417N (G→T) were detected in 33 (91.7%), 34 (94.4%), 1 (2.8%), and 0 (0.0%) samples, respectively, and the incidence rates of the B.1.1.7 (UK) and P.1 (Brazil/Japan) variants were calculated as 33/36 (91.7%) and 1/36 (2.8%), respectively. rRT-PCR and direct sequencing results were concordant in all samples (Table 1).
The incidence rate of the UK variant was very high in the Ulsan area. Recent studies have reported that although the UK variant does not hinder vaccine-induced immunity, it may increase fatality by up to 18% compared with that due to non-UK SARS-CoV-2 variants [9, 10]. These results suggest that UK variant-infected patients need to be managed cautiously. The high incidence of the UK variant in the Ulsan area compared with other areas in Korea [11] can be attributed to the frequent entry of foreigners at the Ulsan trading port, increasing the probability of transmission of variants. Although the incidence rates of the RSA and Brazil/Japan variants were low in our study, considering some limitations, including the small sample size, single institutional nature, and use of direct sequencing rather than whole-genome sequencing for validation, a more comprehensive study is needed to confirm these preliminary results.
In conclusion, the incidence of infection due to the UK variant is very high (91.7%), whereas that due to the RSA and Brazil/Japan variants is low in the Ulsan area of Korea. A large-scale study is required to confirm our results. Powerchek SARS-CoV-2 S-gene Mutation Detection Kit is useful for the rapid screening of SARS-CoV-2 variants.
Notes
AUTHOR CONTRIBUTIONS
Kang H, Kim JH and Lee J were involved in the implementation of SARS-CoV-2 variant screening. Park SH, Kang H, JH and Lee J analyzed the data. Park SH drafted the first submitted manuscript and Jeong J supervised the manuscript preparation and submission. Kim HK, Lim JH and Lee SH contributed to the conception. All authors reviewed the manuscript and provided critical feedback.
REFERENCES
1. Hong KH, Lee SW, Kim TS, Huh HJ, Lee J, Kim SY, et al. 2020; Guidelines for Laboratory Diagnosis of Coronavirus Disease 2019 (COVID-19) in Korea. Ann Lab Med. 40:351–60. DOI: 10.3343/alm.2020.40.5.351. PMID: 32237288. PMCID: PMC7169629.

2. Choudhary S, Sreenivasulu K, Mitra P, Misra S, Sharma P. 2021; Role of Genetic Variants and Gene Expression in the Susceptibility and Severity of COVID-19. Ann Lab Med. 41:129–38. DOI: 10.3343/alm.2021.41.2.129. PMID: 33063674. PMCID: PMC7591285.

3. Hu B, Guo H, Zhou P, Shi Z-L. 2021; Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 19:141–54. DOI: 10.1038/s41579-020-00459-7. PMID: 33024307. PMCID: PMC7537588.

4. Plante JA, Mitchell BM, Plante KS, Debbink K, Weaver SC, Menachery VD. 2021; The variant gambit: COVID-19's next move. Cell Host Microbe. 29:508–15. DOI: 10.1016/j.chom.2021.02.020. PMID: 33789086. PMCID: PMC7919536.

5. Tortorici MA, Walls AC, Lang Y, Wang C, Li Z, Koerhuis D, et al. 2019; Structural basis for human coronavirus attachment to sialic acid receptors. Nat Struct Mol Biol. 26:481–9. DOI: 10.1038/s41594-019-0233-y. PMID: 31160783. PMCID: PMC6554059.

6. Xie X, Liu Y, Liu J, Zhang X, Zou J, Fontes-Garfias CR, et al. 2021; Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K, and N501Y variants by BNT162b2 vaccine-elicited sera. Nat Med. 27:620–1. DOI: 10.1038/s41591-021-01270-4. PMID: 33558724.

7. Starr TN, Greaney AJ, Addetia A, Hannon WW, Choudhary MC, Dingens AS, et al. 2021; Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 371:850–4. DOI: 10.1126/science.abf9302. PMID: 33495308. PMCID: PMC7963219.

8. Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JC, et al. 2020; Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 28:e61312. DOI: 10.7554/eLife.61312.sa2. PMID: 33112236. PMCID: PMC7723407.

9. Zhao S, Lou J, Chong MKC, Cao L, Zheng H, Chen Z, et al. 2021; Inferring the association between the risk of COVID-19 case fatality and N501Y substitution in SARS-CoV-2. Viruses. 13:638. DOI: 10.3390/v13040638. PMID: 33918060. PMCID: PMC8070306.

10. Conti P, Caraffa A, Gallenga CE, Kritas SK, Frydas I, Younes A, et al. 2021; The British variant of the new coronavirus-19 (Sars-Cov-2) should not create a vaccine problem. J Biol Regul Homeost Agents. 35:1–4. DOI: 10.23812/21-3-E. PMID: 33377359.
11. Kim Y, Kim EJ, Lee SW, Kwon D. 2021; Review of the early reports of the epidemiological characteristics of the B.1.1.7 variant of SARS-CoV-2 and its spread worldwide. Osong Public Health Res Perspect. 12:139–48. DOI: 10.24171/j.phrp.2021.0037. PMID: 34218651. PMCID: PMC8256300.

Fig. 1
Examples of rRT-PCR and direct sequencing results of SARS-CoV-2 variants. (A) rRT-PCR results. Amplification curves for N501Y, E484K, K417N, SARS-CoV-2, and the internal control are shown in red, green, blue, black, and purple, respectively. (B) Direct sequencing results for del69-70, K417N, E484K, and N501Y variants.
Abbreviations: rRT-PCR, real-time reverse transcription PCR; N, asparagine; Y, tyrosine; E, glutamate; K, lysine; IC, internal control; UK, United Kingdom; RSA, Republic of South Africa; T, thymine; G, guanine; A, adenine; C, cytosine; U, uracil.
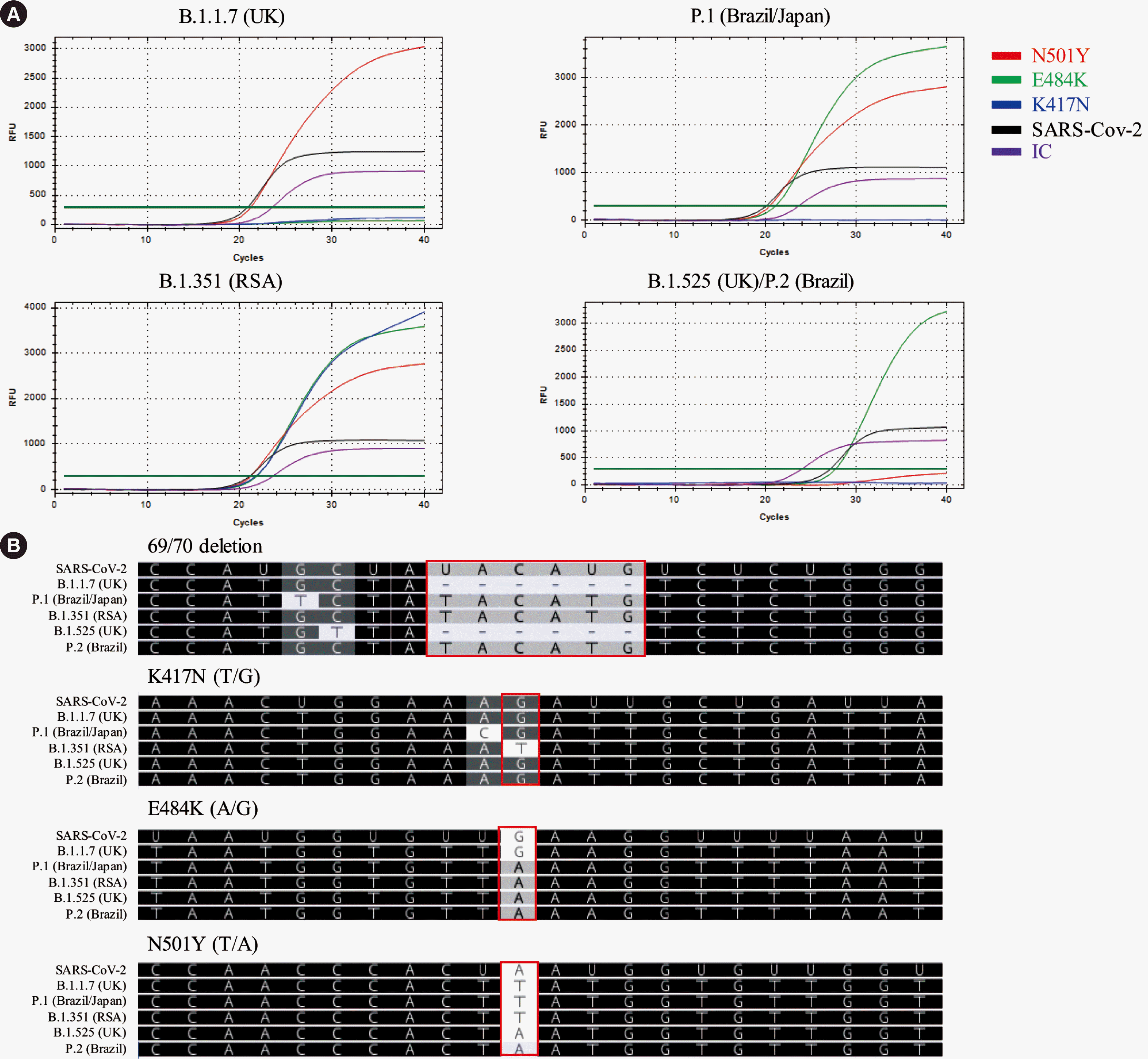
Table 1
Results of rRT-PCR and direct sequencing of 36 SARS-CoV-2 samples




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download