Abstract
Background
We established high-sensitivity cardiac troponin I (hsTnI) 99th percentile upper reference limits (URLs) for the Centaur XPT High-Sensitivity Troponin I assay (Centaur hsTnI; Siemens, Erlangen, Germany) and Atellica IM High-Sensitivity Troponin I assay (Atellica hsTnI; Siemens) and assessed the effect of outlier elimination.
Methods
The reference population comprised 380 men and 387 women, satisfying the strict systematic reference population criteria. After reference population verification by the N-terminal pro-B-type natriuretic peptide (NT-proBNP) assay, 99th percentile URLs for Centaur hsTnI and Atellica hsTnI were calculated before and after outlier elimination.
Results
The 99th percentile URL for Centaur hsTnI was 60.4 (men, 74.7; women, 57.5) ng/L and that for Atellica hsTnI was 59.6 (men, 75.2; women, 55.1) ng/L. After the elimination of 61 (8.0%) outlier samples in Centaur hsTnI and 58 (7.6%) in Atellica hsTnI, the 99th percentile URLs were 13.5 ng/L (men, 15.3 ng/L; women, 11.9 ng/L) and 13.4 ng/L (men, 15.5 ng/L; women, 12.9 ng/L), respectively, significantly lower than those before outlier elimination. The CVs at the 99th percentile URLs were 5.2% and 3.5%, respectively. The measurable fractions among the reference population were 91.5% and 93.4%, respectively. Performance evaluation of Atellica B-type natriuretic peptide (BNP), Atellica NT-proBNP, Centaur hsTnI, and Atellica hsTnI showed outstanding results.
Conclusions
The Korean hsTnI 99th percentile URLs calculated in this study were significantly lower after outlier elimination than before. Centaur hsTnI and Atellica hsTnI meet the “Guideline acceptable” and “Level 3 (second generation, high sensitivity)” requirements, satisfying international standards.
Troponin (Tn), a structural protein, is a component of actin-myosin fibers and is involved in calcium-mediated muscle contraction. Among Tns, cardiac (c)TnT and cTnI are often used to diagnose and monitor cardiac diseases as they are specific to cardiac muscles [1, 2]. According to the European Society of Cardiology, American College of Cardiology, American Heart Association, and World Heart Federation Task Force Fourth Universal Definitions of Myocardial Injury and Myocardial Infarction, myocardial infarction can be diagnosed when cTn values are above the 99th percentile of the upper reference limit (URL) values determined in healthy individuals and when at least one cardiac symptom specified in the guidelines occurs [2]. Hence, for rapid and accurate diagnosis of myocardial infarction, it is crucial to measure cTn values accurately and to establish a reference range for cTn based on a strictly selected healthy reference population [3, 4].
Multiple studies have reported that the 99th percentile URLs used as a cTn reference range in diagnosing myocardial infarction vary across demographics, especially considering the size and clinical characteristics of the reference population [5-9]. In 2014, Sandoval and Apple [10] reported strict standards for selecting a reference population to establish 99th percentile URLs for cTn. According to these standards, at least 300 men and 300 women should be recruited. The American Association for Clinical Chemistry (AACC) and International Federation of Clinical Chemistry and Laboratory Medicine (IFCC) guidelines suggest the same criteria for the reference population size required to define the 99th percentile URL for each sex [11]. However, Hickman, et al. [12] showed that outlier elimination can dramatically alter cTn 99th percentile URLs. Thus, incorrect outlier evaluation may significantly increase 99th percentile URLs [6, 9, 12, 13].
Recently, the ADVIA Centaur High-Sensitivity Troponin I assay (Centaur hsTnI) (Siemens, Erlangen, Germany), which is a fully automated version of the Centaur XP TnI-Ultra assay (Centaur TnI; Siemens), and the Atellica IM High-Sensitivity Troponin I assay (Atellica hsTnI; Siemens) were introduced in clinical laboratories. The Centaur TnI and both hsTnI assays are chemiluminescence immunoassays. However, the hsTnI assays newly employ trisulfopropyl acridinium ester, a strong anionic label with very low non-specific binding and the same high light yield that confers the hsTnI assays a 10-fold improvement in low-end precision sensitivity. Additionally, the hsTnI assays are robust, with a high tolerance for common interferents, such as biotin, hemolysis, and alkaline phosphatase. Further, they show an optimal precision of >90% across a measurable range. The two hsTnI assays have subtle differences. The time to first result is 18 and 10 minutes for Centaur hsTnI and Atellica hsTnI, respectively. In addition, the 99th percentile value (for men and women combined and for serum, according to the package insert) is 46.8 ng/L and 45.4 ng/L for Centaur hsTnI and Atellica hsTnI, respectively. Both assays analyze the full cardiac panel, including high-sensitivity TnI (hsTnI), B-type natriuretic peptide (BNP), N-terminal pro-brain BNP (NT-proBNP), myoglobulin, and creatine kinase myocardial band, in a single platform.
We calculated 99th percentile URLs for cTnI for Centaur hsTnI and Atellica hsTnI in a healthy Korean population of sufficient size, meeting the selection criteria suggested by Sandoval and Apple and the AACC and IFCC guidelines [10, 11], to determine the effect of outlier elimination on the 99th percentile URLs. Additionally, we evaluated the performance of Centaur hsTnI, Atellica hsTnI, Atellica IM BNP, and Atellica IM NT-proBNP, all of which can be used to assess cardiac function.
This retrospective observational study was reviewed and approved by the Institutional Review Board (IRB) of Asan Medical Center, Seoul, Korea (approval number.: 2018-0731). As we used residual serum samples that had been used for regular laboratory assays and were completely anonymized, the requirement for informed consent was waived by the IRB.
Residual serum samples bottled in BD Vacutainer Plus Plastic Serum Tubes (BD, Franklin Lakes, NJ, USA) from subjects who visited Asan Medical Center for general health checkups between October 10, 2019 and February 25, 2020 were collected. Within 24 hours after collection, the samples were divided into four 250-μL aliquots in Eppendorf tubes (Eppendorf North America, Enfield, CT, USA) and frozen at –70°C until assaying. At the end of the sample collection, all frozen samples were thawed, vortexed, and centrifuged for analysis. These samples were used to evaluate cTnI 99th percentile URLs. The inclusion criteria for reference population selection and the study flowchart are presented in Fig. 1.
To assess “clinical history for known cardiovascular disease and medication usage,” we reviewed the subjects’ checkup questionnaire data, electronic medical records, and laboratory test results at the time of recruitment to screen those with active or historical cardiac diseases, including acute coronary syndromes, congestive heart failure, myocarditis, ventricular tachyarrhythmias, and percutaneous coronary intervention or cardiac medication use. All subjects also underwent NT-proBNP measurement, which is a criterion for selecting the reference population, to screen out those with current or historical asymptomatic cardiac diseases, as these can cause outliers and may affect the calculation of the 99th percentile URL [14, 15].
The NT-proBNP cutoff was set at the age- and sex-specific values provided by the manufacturer (package insert of the Elecsys NT-proBNP assay, Roche, Basel, Switzerland) [18-54 years: 143.0 (men) or 208.4 (women) ng/L; 55–64 years: 650.8 (men) or 912.2 (women) ng/L; 65–74 years: 512.0 (men) or 533.4 (women) ng/L; ≥75 years: 603.6 (men) or 665.0 (women) ng/L]. To include at least 300 men and 300 women in the calculation of the cTnI 99th percentile URL, the subjects were divided into the following age groups: <30, 30–39, 40–49, 50–59, 60–69, and ≥70 years. The subjects were further divided into subgroups based on age ≤55 years or >55 years to compare the effect of age on TnI 99th percentile URLs.
Precision was evaluated according to the CLSI guidelines EP5-A3 [16]. Low, medium, and high levels of quality control (QC) materials (Liquichek Cardiac Markers Plus Control, lot numbers 99562, 99563, and 99565; Bio-Rad, Hercules, CA, USA) were used for Atellica IM BNP, Atellica IM NT-pro BNP, Centaur hsTnI, and Atellica hsTnI. Each QC standard was analyzed in duplicate and twice a day (with a ≥2-hour interval between analyses) for 20 days.
Linearity was evaluated according to the CLSI guidelines EP6-A [17], using Atellica IM BNP Master Curve Material, Atellica IM PBNP Master Curve Material, Atellica IM TNIH Master Curve Material, and ADVIA Centaur TNIH Master Curve Material (all from Siemens). Each material was assayed four times. The data were subjected to polynomial regression analysis, and the deviation from linearity was calculated. The allowable nonlinearity was set at 27.9% for hsTnI and 13% for BNP and ProBNP based on the total allowable error of the Desirable Biological Variation Database specifications available on the Westgard website [18].
Method comparison was conducted according to the CLSI guidelines EP9-A2 [19]. For BNP and hsTnI, Centaur XPT performance was compared with that of Atellica IM. For NT-proBNP, Cobas C8000 (Roche) was used in comparison with Atellica IM. To compare cTnI and hsTnI, Centaur XPT was used. Forty residual serum samples were analyzed separately for each of the four analytes. Data were analyzed using Deming regression, and the slope, intercept, and correlation coefficients (R values) were calculated. R2 ≥0.95 indicated a linear correlation between the two reagents over the comparison experiment range.
We obtained whole blood samples from two healthy volunteers with informed consent. Serum was separated immediately after blood collection and stored at –70°C. The serum samples were thawed right before use and spiked with residual serum samples that showed high cTnI values to prepare serum pools with TnI values of 26.5–106.3 ng/L. Using the multilevel pools, we generated precision profile plots. Samples with six TnI values spanning the 99th percentile URLs were prepared. The TnI value of each sample was measured 80 times using Centaur hsTnI and Atellica hsTnI. The CVs at each TnI value for each assay were then analyzed according to the CLSI guidelines EP5-A3 [16]. Based on the CVs of the six serum pools, we generated precision profile plots using the Variance Function Program (version 15.0; W. A. Sadler, Christchurch, New Zealand) and calculated the CVs at the 99th percentile URLs for Centaur hsTnI and Atellica hsTnI.
The presence of outliers in data is a major problem and strongly affects 99th percentile URLs [8, 9, 11, 20, 21]. We determined the 99th percentile URLs using the total samples before and after outlier elimination by a one-tailed non-parametric method with 3,000 bootstrap replications and a 95% confidence interval (CI) in SPSS (version 18.0.0; IBM, Armonk, NY, USA). Outliers were identified using Tukey’s 3-interquartile range (IQR) test using the following formula: outlier hsTnI value >Q3+3 IQR, where Q3 and IQR are the third quartile and interquartile range (Q3–Q1) of the hsTnI distribution, respectively [9, 12]. We eliminated outliers manually. Since hsTnI values are higher in older subjects, especially at the reference age of 55 years in the general population, we stratified the reference population by age (≤55 years and >55 years) and determined the 99th percentile URLs for each group [8, 9].
As hsTnI values were not normally distributed, sex- and age-related differences were evaluated using the Mann-Whitney and Kruskal-Wallis tests. hsTnI values are represented as the median and 95% CI [22]. Statistical significance was set at P<0.05.
For general statistical analysis, including the four analyte performance evaluations, EP Evaluator Release 9 (Data Innovations, Colchester, VT, USA) and Microsoft Excel 2016 (Microsoft, Redmond, WA, USA) were used. To calculate the 99th percentile URLs and CVs, Variance Function Program version 15.0 and SPSS version 18.0.0. (IBM) were used.
The results of precision analyses are presented in Supplemental Data Table S1. For all assays, the total CV was 2.0%–4.1%. The CVs at the 99th percentile URLs were both <10%, which satisfied the criteria for hsTn assays according to the AACC and IFCC guidelines [11].
The results of linearity evaluation are presented in Supplemental Data Fig. S1. Linearity was observed throughout the estimated analytical range for all analytes. R2 ≥0.99 was observed for all analytes, with R2=0.99 for Centaur hsTnI at 0.0–19,318.6 ng/L, R2=0.99 for Atellica hsTnI at 1.9–18,884.9 ng/L, R2=0.99 for Atellica IM BNP at 0.9–4,000.1 ng/L, and R2=0.99 for Atellica IM NT-proBNP at 0.6–29,593.9 ng/L. The percentage recovery ranged from 90% (BNP, low value) to 101% (NT-proBNP, high value).
Scatter and bias plots of the results of method comparison for BNP using Atellica IM and Centaur XPT, for NT-proBNP using Atellica IM and Roche Cobas 8000, for hsTnI using Atellica IM and Centaur XPT, and for cTnI and hsTnI using Centaur XPT for both are presented in Supplemental Data Fig. S2. For all analytes, the results showed strong correlations (R2 ≥0.95). Specifically, the correlation equation for BNP between Atellica IM and Centaur XPT was y=0.98 x+0.69 (R2=0.99), showing a strong correlation between the two assays. The correlation coefficient for NT-proBNP between Atellica IM and Roche Cobas 8000 was strong (R2=0.99), with a correlation equation of y=1.22 x+23.35. The correlation equation and coefficient for hsTnI between Atellica hsTnI and Centaur hsTnI were strong (y=0.99 x−11.99, R2=0.99). cTnI values using Centaur XPT and hsTnI values using Centaur XPT showed strong correlation (R2=0.96). The correlation equation slope was 0.853, indicating a proportional difference between the two assays. Hence, it is essential for institutions to re-establish 99th percentile URLs when replacing Centaur TnI with Centaur hsTnI.
After considering the NT-proBNP assay results, five men and three women showed higher values than the aforementioned cutoff values and were excluded from the reference population. We obtained 380 samples from men and 387 samples from women before outlier elimination. Fig. 2 shows the TnI value distribution in the reference population in men and women before outlier elimination.
The hsTnI value distributions according to age and sex in the reference population are shown in Figs. 3 and 4. Overall, hsTnI values were higher in men than in women within the same age group and tended to increase with age.
The 99th percentile URLs are listed in Table 1. The 99th percentile URLs of Centaur hsTnI and Atellica hsTnI before outlier elimination were 60.4 and 59.6 ng/L, respectively. After outlier elimination, which accounted for 6.6%–10.1% of each subgroup, the number of subjects in the reference population decreased to 349 men and 357 women for Centaur hsTnI and to 352 men and 357 women for Atellica hsTnI, and the 99th percentile URLs significantly decreased to 13.5 and 13.4 ng/L for Centaur hsTnI and Atellica hsTnI, respectively.
When comparing men and women stratified according to age (≤55 years vs. >55 years), the hsTnI 99th percentile URLs of Centaur hsTnI and Atellica hsTnI were significantly higher in men aged >55 years than in women aged ≤55 years [8, 9] (P<0.001 for both; Fig. 4). The proportion of measurable samples above the limit of detection (LoD) was 91.5% and 93.4% for Centaur hsTnI and Atellica hsTnI, respectively (Table 2).
Various studies have indicated that TnI 99th percentile URLs for Asians are lower than those for Caucasians [6, 7, 23-26]. Surprisingly, the 99th percentile URLs obtained in our study before outlier elimination were higher than those previously reported and those provided by the manufacturer (Siemens). Furthermore, they were higher than the 99th percentile URLs of 20.0 ng/L (men) and 11.6 ng/L (women) for Siemens ADVIA Centaur TnI in Koreans [5, 6].
As shown in Figs. 3 and 4, the average hsTnI values increased with age, particularly at ages ≥55 years. Additionally, hsTnI values were significantly higher in men than in women within the same age groups for both assays. The Mann-Whitney test for comparison of hsTnI values in men and women showed P<0.031 for both Centaur hsTnI and Atellica hsTnI. This result may be explained by the possibility of remaining outliers that were not eliminated during statistical analysis, especially in the >55 years age group for both sexes.
Although the manufacturers did not provide detailed standards for the recruitment of the reference population in which 99th percentile URLs are to be determined, the proportion of subjects aged ≥50 years for the 99th percentile URLs established by the manufacturer (Siemens) was 46%. In our reference population, subjects aged ≥50 years accounted for 78.7%. The fact that the 99th percentile URLs in our study were lower than those provided by the manufacturer can be explained by the more stringent selection criteria used in our study than those used in the manufacturer’s study.
The 99th percentile URLs in men and women were 15.3 and 11.9 ng/L and 15.5 and 12.9 ng/L for Centaur hsTnI and Atellica hsTnI, respectively. Kim, et al. [26] have reported that the 99th percentile URLs in men and women for Beckman Coulter Access hsTnI were 11.3 and 9.5 ng/L, respectively, in the Korean population. Apple, et al. [27] reported that hsTnI 99th percentile URLs were 35–43 ng/L for Centaur hsTnI and 30–43 ng/L for Atellica, according to age in a United States population. In a mixed-sex population of 2,010 apparently healthy individuals from Italy, the 99th percentile URL value for Centaur hsTnI was 47.30 ng/L [28]. Based on the results of these and our studies, we conclude that hsTnI 99th percentile URLs are higher for Caucasians than for East Asians and specifically, Koreans.
The hsTnI 99th percentile URLs are significantly higher in men than in women across all ages [23, 29]. In our previous study, 99th percentile URLs calculated for Centaur TnI in Korean men and women were 20.0 and 11.6 ng/L, respectively [6]. As hsTnI 99th percentile URLs are higher in men than in women, it is recommended that different hsTnI 99th percentile URLs are used for men and women. Furthermore, 99th percentile URLs are higher in the older population than in the younger population. Thus, establishing guidelines for age distribution in the reference population used for calculating hsTnI 99th percentile URLs should be considered.
We previously reported a CV of 43.0% at the 99th percentile URL using Centaur TnI, the predecessor of Centaur hsTnI [6]. In contrast, in this study, Centaur hsTnI and Atellica hsTnI showed CVs of 5.2% and 3.5%, respectively, at the 99th percentile URLs, indicating an improvement in precision. Applying the criteria suggested by Apple [30], the CVs at the 99th percentile URLs for Centaur hsTnI and Atellica hsTnI were ≤10%, which is clinically considered “Guideline acceptable.” The measurable normal values (%), defined as the proportions of specimens with values above the LoD, were 91.5% and 93.4% for Centaur hsTnI and Atellica hsTnI, respectively. Both systems were classified as level 3 (second generation, high sensitivity) in a scorecard designation [30]. Current AACC and IFCC guidelines state that hsTn assays must have an analytical imprecision of <10% CV at the 99th percentile URL in a healthy population and should be able to measure cTn above the LoD in 50% of the healthy population, ideally 95%. Both hsTnI assays can be defined as highly sensitive according to the AACC and IFCC criteria [11].
We performed the NT-proBNP assay in the reference population to screen for subjects with asymptomatic cardiac diseases, which can be considered a strength of the current study. However, we are aware that NT-proBNP assays cannot screen cardiac patients perfectly.
This study had some limitations. First, the reference population used to determine hsTnI 99th percentile URLs was smaller than those used in previous similar studies [8, 9]. The number of outliers was relatively high, which reduced the size of the final reference population used to calculate the 99th percentile URLs. It was difficult to include subjects in the reference population as our study was a single-center study, as opposed to multicenter or multinational studies. Second, in the comparison study, we did not use samples covering a wide range of values. In the comparison between Centaur TnI and Centaur hsTnI, samples with values ranging from 0.0 ng/L to 23.5 ng/L were used, which did not cover the entire analytical measurement range of the assays. In comparing NT-proBNP values between the Cobas C8000 and Atellica IM systems, there was a gap between 4,100 ng/L and 15,700 ng/L, and thus, our results do not reflect actual assay results obtained in clinical laboratories. Third, we used frozen serum instead of fresh serum for analysis, which may have influenced the study results. Finally, while recruiting the reference population, we did not use imaging modalities, such as cardiac sonography or electrocardiography. Asymptomatic cardiac patients may have been missed, which may have increased the 99th percentile URLs.
Currently, hsTnI and hsTnT values are critical in diagnosing cardiac diseases, especially, acute myocardial infarction. These values are interpreted using various methods, and quality control of hsTnI and hsTnT is becoming essential to obtain accurate hsTnI and hsTnT values [31, 32]. Our study highlighted the importance of outlier elimination in determining precise 99th percentile URL values.
Considering the clinical significance of hsTnI 99th percentile URLs, determining 99th percentile URLs using a strictly selected population and a reliable and standardized method, including proper outlier elimination, is important. To the best of our knowledge, this is the first study to report an outlier elimination effect in calculating hsTnI 99th percentile URL values, especially in a Korean population. When introducing a new hsTnI assay system, the 99th percentile URLs should be recalculated for clinical application as the Centaur hsTnI assay was not harmonized with the Centaur TnI system according to our results.
ACKNOWLEDGMENTS
We thank Editage (www.editage.co.kr) for editing and reviewing the English language translation of this manuscript. The authors also wish to thank Young Kim and Siemens Healthineers for supporting this project and providing technical assistance.
Notes
REFERENCES
1. Bodor GS, Porterfield D, Voss EM, Smith S, Apple FS. 1995; Cardiac troponin-I is not expressed in fetal and healthy or diseased adult human skeletal muscle tissue. Clin Chem. 41:1710–5. DOI: 10.1093/clinchem/41.12.1710. PMID: 7497610.

2. Thygesen K, Alpert JS, Jaffe AS, Chaitman BR, Bax JJ, Morrow DA, et al. 2018; Fourth universal definition of myocardial infarction (2018). Circulation. 138:e618–51. DOI: 10.1161/CIR.0000000000000617. PMID: 30571511.

3. Reichlin T, Hochholzer W, Bassetti S, Steuer S, Stelzig C, Hartwiger S, et al. 2009; Early diagnosis of myocardial infarction with sensitive cardiac troponin assays. N Engl J Med. 361:858–67. DOI: 10.1056/NEJMoa0900428. PMID: 19710484.

4. Mahajan VS, Jarolim P. 2011; How to interpret elevated cardiac troponin levels. Circulation. 124:2350–4. DOI: 10.1161/CIRCULATIONAHA.111.023697. PMID: 22105197.

5. Collinson PO, Heung YM, Gaze D, Boa F, Senior R, Christenson R, et al. 2012; Influence of population selection on the 99th percentile reference value for cardiac troponin assays. Clin Chem. 58:219–25. DOI: 10.1373/clinchem.2011.171082. PMID: 22100808.

6. Ko DH, Jeong TD, Cho EJ, Lim J, Ji M, Lee K, et al. 2017; The 99th percentile values of six cardiac troponin assays established for a reference population using strict selection criteria. Clin Chim Acta. 464:1–5. DOI: 10.1016/j.cca.2016.11.004. PMID: 27823950.

7. Di Pietro M, Dipalo M, Rocchi MBL, Musa R, Avanzini P, Gnocchi C, et al. 2019; Assessment of Access hsTnI 99th percentiles upper reference limits following IFCC recommendations. Clin Chim Acta. 492:26–8. DOI: 10.1016/j.cca.2019.01.028. PMID: 30711523.

8. Clerico A, Zaninotto M, Ripoli A, Masotti S, Prontera C, Passino C, et al. 2017; The 99th percentile of reference population for cTnI and cTnT assay: methodology, pathophysiology and clinical implications. Clin Chem Lab Med. 55:1634–51. DOI: 10.1515/cclm-2016-0933. PMID: 28599373.

9. Clerico A, Ripoli A, Zaninotto M, Masotti S, Musetti V, Ciaccio M, et al. 2019; Head-to-head comparison of plasma cTnI concentration values measured with three high-sensitivity methods in a large Italian population of healthy volunteers and patients admitted to emergency department with acute coronary syndrome: A multi-center study. Clin Chim Acta. 496:25–34. DOI: 10.1016/j.cca.2019.06.012. PMID: 31201817.

10. Sandoval Y, Apple FS. 2014; The global need to define normality: the 99th percentile value of cardiac troponin. Clin Chem. 60:455–62. DOI: 10.1373/clinchem.2013.211706. PMID: 24115136.

11. Wu AHB, Christenson RH, Greene DN, Jaffe AS, Kavsak PA, Ordonez-Llanos J, et al. 2018; Clinical Laboratory practice recommendations for the use of cardiac troponin in acute coronary syndrome: expert opinion from the Academy of the American Association for Clinical Chemistry and the Task Force on Clinical Applications of Cardiac Bio-markers of the International Federation of Clinical Chemistry and Laboratory Medicine. Clin Chem. 64:645–55. DOI: 10.1373/clinchem.2017.277186. PMID: 29343532.

12. Hickman PE, Koerbin G, Potter JM, Abhayaratna WP. 2017; Statistical considerations for determining high-sensitivity cardiac troponin reference intervals. Clin Biochem. 50:502–5. DOI: 10.1016/j.clinbiochem.2017.02.022. PMID: 28263716.

13. Eggers KM, Apple FS, Lind L, Lindahl B. 2016; The applied statistical approach highly influences the 99th percentile of cardiac troponin I. Clin Biochem. 49:1109–12. DOI: 10.1016/j.clinbiochem.2016.08.012. PMID: 27556285.

14. Clerico A, Zaninotto M, Passino C, Aspromonte N, Piepoli MF, Migliardi M, et al. 2020; Evidence on clinical relevance of cardiovascular risk evaluation in the general population using cardio-specific biomarkers. Clin Chem Lab Med. 59:79–90. DOI: 10.1515/cclm-2020-0310. PMID: 32692693.

15. Farmakis D, Mueller C, Apple FS. 2020; High-sensitivity cardiac troponin assays for cardiovascular risk stratification in the general population. Eur Heart J. 41:4050–6. DOI: 10.1093/eurheartj/ehaa083. PMID: 32077940.

16. CLSI. 2004. Evaluation of precision performance of quantitative measurement methods: approved guideline. EP5-A2. 2nd ed. Clinical and Laboratory Standards Institute;Wayne, PA:
17. CLSI. 2003. Evaluation of the linearity of quantitative measurement procedures: A statistical approach: approved guideline. EP6-A. Clinical and Laboratory Standards Institute;Wayne, PA:
18. Ricos C, Alvarez V, Cava F, Garcia-Lario JV, Hernandez A, Jimenez CV, et al. Desirable Biological Variation Database specifications: Desirable specifications for total error, imprecision, and bias, derived from intra- and inter-individual biologic variation. https://www.westgard.com/biodatabase1.htm. Updated in 2014.
19. CLSI. 2002. Method comparison and bias estimation using patient samples: approved guidelines. EP9-A2. 2nd ed. Clinical and Laboratory Standards Institute;Wayne, PA:
20. Franzini M, Lorenzoni V, Masotti S, Prontera C, Chiappino D, Latta DD, et al. 2015; The calculation of the cardiac troponin T 99th percentile of the reference population is affected by age, gender, and population selection: a multicenter study in Italy. Clin Chim Acta. 438:376–81. DOI: 10.1016/j.cca.2014.09.010. PMID: 25239669.

21. Hickman PE, Potter JM, Aroney C, Koerbin G, Southcott E, Wu AH, et al. 2010; Cardiac troponin may be released by ischemia alone, without necrosis. Clin Chim Acta. 411:318–23. DOI: 10.1016/j.cca.2009.12.009. PMID: 20036224.

22. Tukey J. Exploratory data analysis, 1977. http://www.ru.ac.bd/wp-content/uploads/sites/25/2019/03/102_05_01_Tukey-Exploratory-Data-Analysis-1977.pdf.
23. Clerico A, Masotti S, Musetti V, Ripoli A, Aloe R, Di Pietro M, et al. 2019; Evaluation of 99th percentile and reference change values of the hs-cTnI method using ADVIA Centaur XPT platform: A multicenter study. Clin Chim Acta. 495:161–6. DOI: 10.1016/j.cca.2019.04.056. PMID: 30978328.

24. Ji M, Moon HW, Hur M, Yun YM. 2016; Determination of high-sensitivity cardiac troponin I 99th percentile upper reference limits in a healthy Korean population. Clin Biochem. 49:756–61. DOI: 10.1016/j.clinbiochem.2016.01.027. PMID: 27067595.

25. Moon SY, Kim N, Lee SM, Yu S, Jun KR, Kim HH. 2019; 99th percentile upper reference limit of AccuTnI+3 in a Korean reference population: a multicenter study using fresh serum. Clin Chem Lab Med. 57:e282–4. DOI: 10.1515/cclm-2019-0189. PMID: 31054248.

26. Kim S, Yoo SJ, Kim J. 2020; Evaluation of the new Beckman Coulter Access hsTnI: 99th percentile upper reference limits according to age and sex in the Korean population. Clin Biochem. 79:48–53. DOI: 10.1016/j.clinbiochem.2020.02.005. PMID: 32059836.

27. Apple FS, Wu AHB, Sandoval Y, Sexter A, Love SA, Myers G, et al. 2020; Sex-Specific 99th percentile upper reference limits for high sensitivity cardiac troponin assays derived using a universal sample bank. Clin Chem. 66:434–44. DOI: 10.1093/clinchem/hvz029. PMID: 32109298.

28. Musetti V, Masotti S, Prontera C, Storti S, Ndreu R, Zucchelli GC, et al. 2018; Evaluation of the analytical performance of a new ADVIA immunoassay using the Centaur XPT platform system for the measurement of cardiac troponin I. Clin Chem Lab Med. 56:e229–31. DOI: 10.1515/cclm-2018-0054. PMID: 29679526.

29. Petersmann A, Ittermann T, Fries C, Lubenow N, Kohlmann T, Kallner A, et al. 2013; Comparison of the 99th percentiles of three troponin I assays in a large reference population. Clin Chem Lab Med. 51:2181–6. DOI: 10.1515/cclm-2013-0113. PMID: 23846150.

30. Apple FS. 2009; A new season for cardiac troponin assays: it's time to keep a scorecard. Clin Chem. 55:1303–6. DOI: 10.1373/clinchem.2009.128363. PMID: 19478023.

31. Kim JW, Kim H, Yun YM, Lee KR, Kim HJ. 2020; Absolute change in high-sensitivity cardiac troponin I at three hours after presentation is useful for diagnosing acute myocardial infarction in the emergency department. Ann Lab Med. 40:474–80. DOI: 10.3343/alm.2020.40.6.474. PMID: 32539303. PMCID: PMC7295960.

32. Li T, Cao S, Wang Y, Xiong Y, He Y, Ke P, et al. 2021; Moving rate of positive patient results as a quality control tool for high-sensitivity cardiac troponin T assays. Ann Lab Med. 41:51–9. DOI: 10.3343/alm.2021.41.1.51. PMID: 32829579. PMCID: PMC7443519.

Fig. 1
Flow diagram showing the reference population used, process of outlier elimination, and the final 99th percentile URLs calculated for Centaur hsTnI and Atellica hsTnI.
Abbreviations: eGFR, estimated glomerular filtration rate; CKD-EPI, Chronic Kidney Disease Epidemiology Collaboration; HbA1c, hemoglobin A1c; NT-proBNP, N-terminal pro-B-type natriuretic peptide; URL, upper reference limit; hsTnI, high-sensitivity troponin I; IQR, interquartile range.

Fig. 2
Distribution of TnI values in the reference population, which was selected according to the strict selection criteria suggested by Sandoval, et al. [10], without outlier elimination. Arrows indicate subjects with outlying TnI values according to Tukey’s 3-IQR method. (A) Men, (B) women.
Abbreviations: TnI, troponin I; IQR, interquartile range.
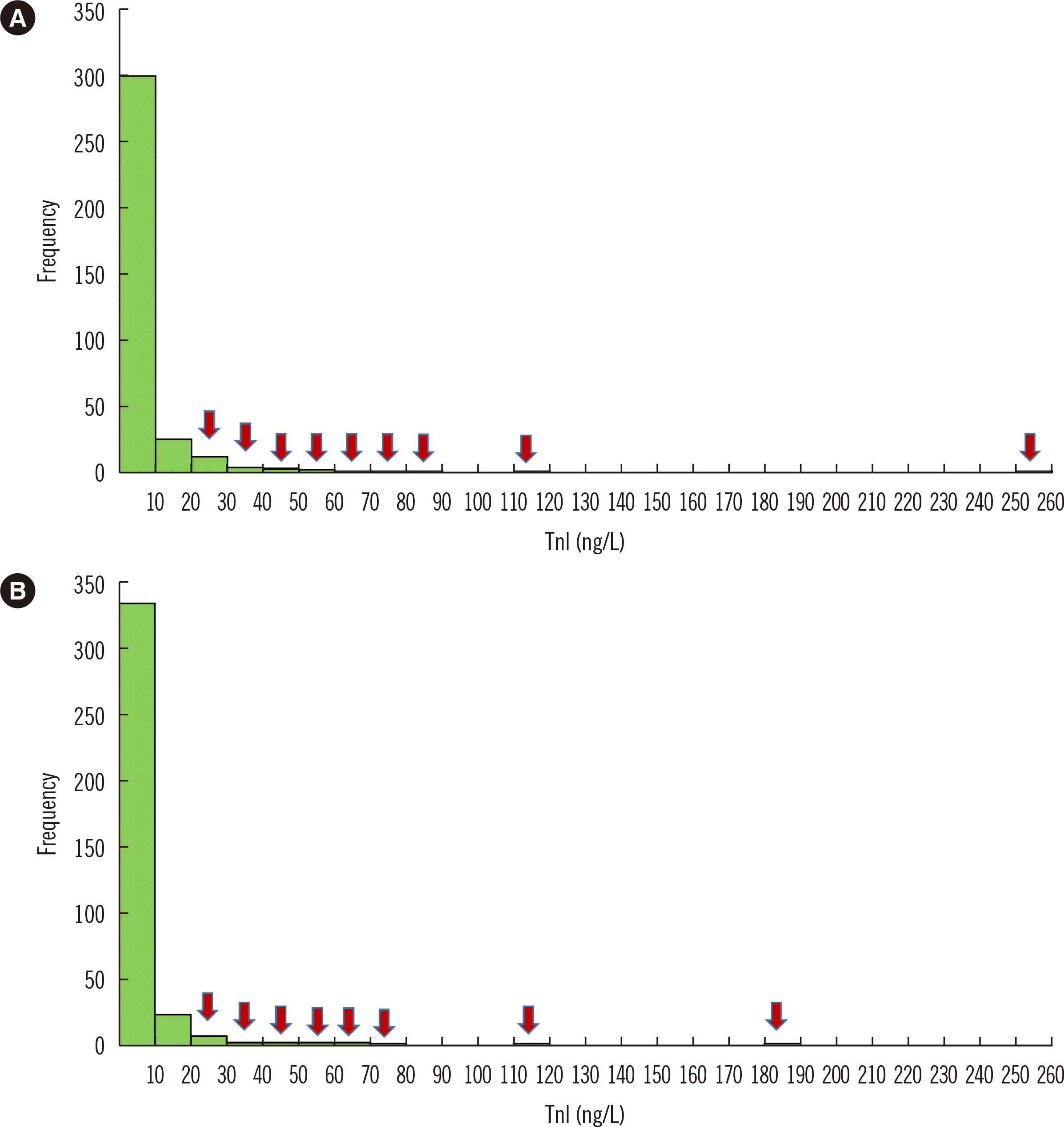
Fig. 3
Distribution of hsTnI values in men and women according to age and sex. The box indicates the 25th and 75th percentiles; the black line corresponds to the median value. (A) Centaur hsTnI, (B) Atellica hsTnI.
Abbreviation: hsTnI, high-sensitivity troponin I.
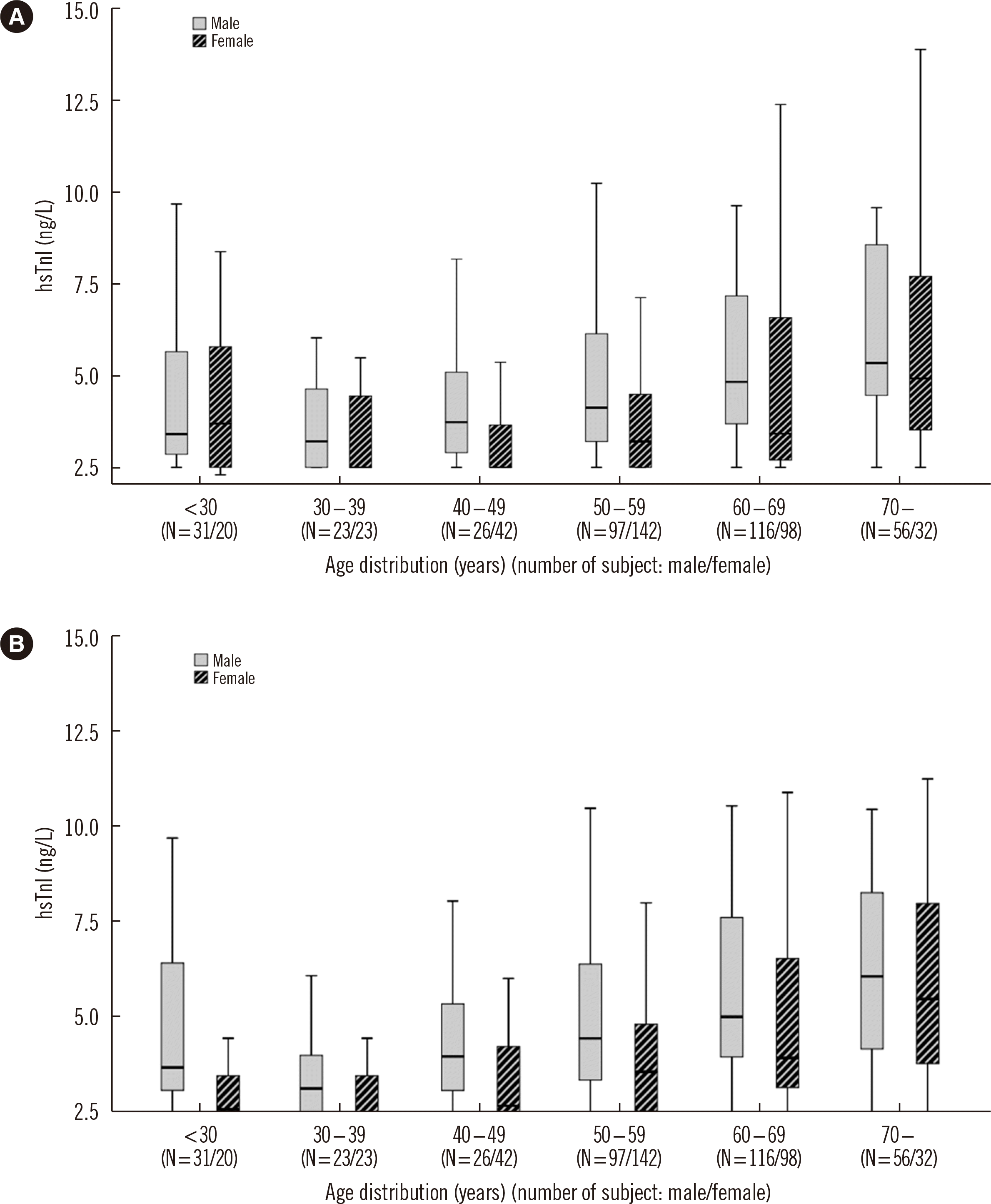
Fig. 4
hsTnI values in four subpopulations according to sex and age (≤55 or >55 years). The box indicates the 25th and 75th percentiles, and the horizontal black line corresponds to the median value. The differences among the groups were statistically significant (P<0.001, Kruskal–Wallis test), with increasing tendencies in the following order: (A) Centaur hsTnI, ≤55 years, women (2.5, 2.5–3.1), all ages, women (3.2, 2.5–4.5), ≤55 years, men (3.7, 2.8–4.7), >55 years, women (3.7, 2.8–5.6), all ages, men (4.2, 3.3–5.7), >55 years, men (4.6, 3.6–6.2) (B) Atellica hsTnI, ≤55 years, women (2.5, 2.5–3.3), all ages, women (3.5, 2.5–4.7), ≤55 years, men (3.9, 3.1–5.1), >55 years, women (4.0, 3.3–6.0), all ages, men (4.2, 3.3–5.9), and >55 years, men (4.9, 3.7–6.6). Data are expressed as (median, IQR) ng/L.
Abbreviations: hsTnI, high-sensitivity troponin I; IQR, interquartile range.
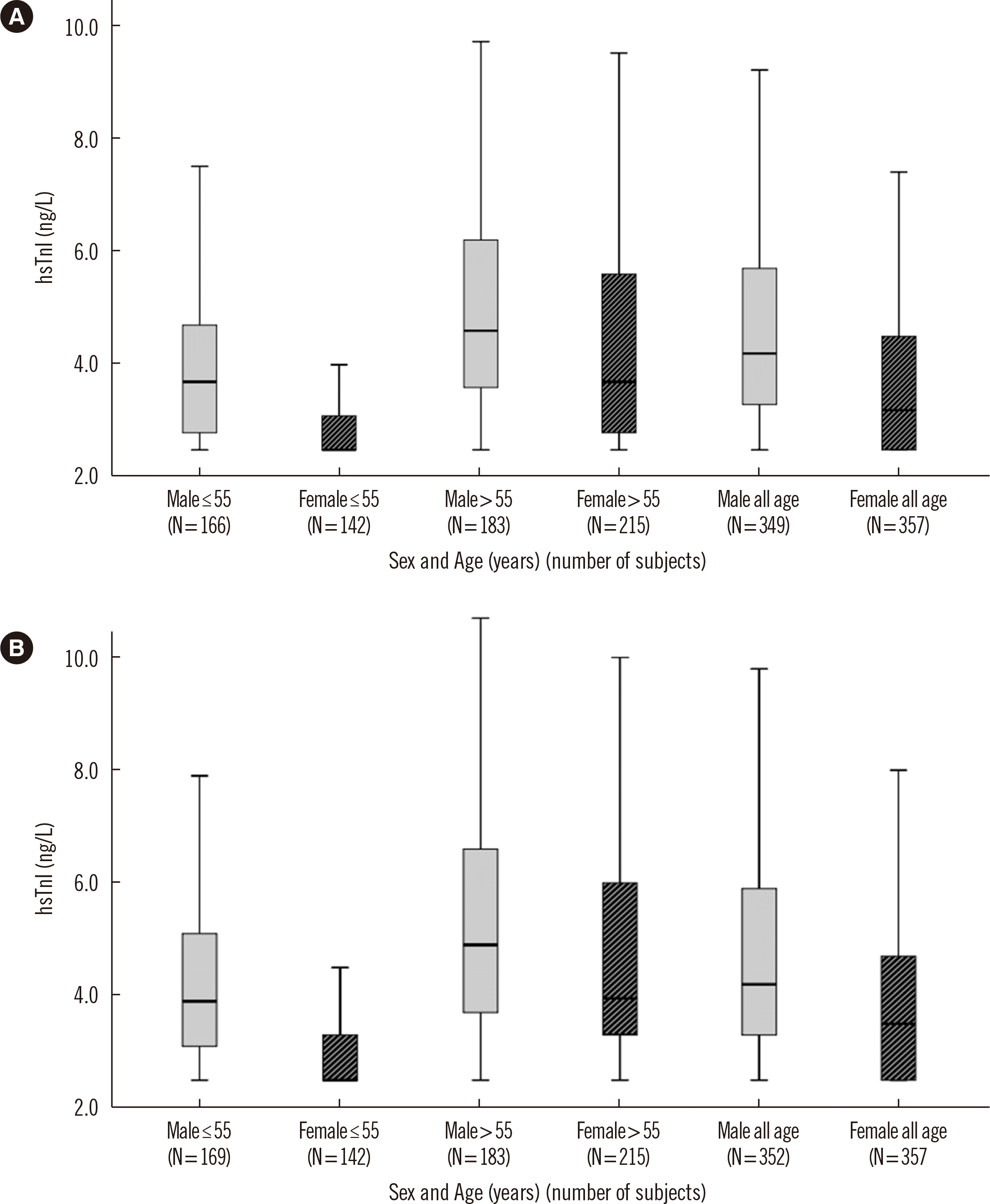
Table 1
hsTnI 99th percentile URLs according to sex, age group, and before and after outlier elimination.
Table 2
hsTnI 99th percentile URLs (ng/L), CV% at the URL, and proportion (%) of measurable samples above the LoD for both assays
URLs were determined based on AACC and AFCC guidelines. Assay designation was performed according to the proportion (%) of measurable samples (Apple, 2009 [30]).




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download