INTRODUCTION
MATERIALS AND METHODS
Preparation of SF
Cell culture and treatment
Cell viability assay
Measurement of nitric oxide (NO), prostaglandin E2 (PGE2), and cytokines
Protein isolation and Western blot analysis
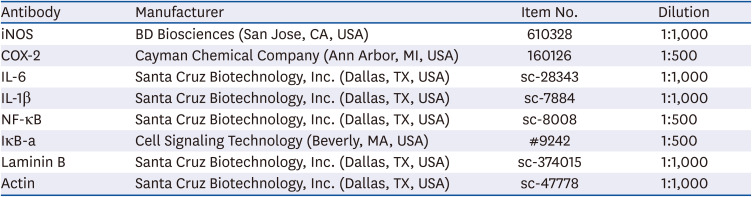
Table 1
List of antibodies used for western blot analysis in the present study
Reverse transcription-polymerase chain reaction (RT-PCR) assay
Immunofluorescence for NF-κB
Measurement of ROS levels
Zebrafish maintenance and PM2.5 treatment
NO and ROS staining in zebrafish larvae
Statistical analysis
RESULTS
Effect of SF on the proliferation of RAW 264.7 macrophages
SF inhibits PM2.5-induced NO and PGE2 production in RAW 264.7 macrophages
 | Fig. 1Effect of SF on the production pro-inflammatory mediators and cytokines in PM2.5-stimulated RAW 264.7 macrophages. Cells were treated with the indicated concentrations of SF for 1 h and then stimulated with 50 µg/mL PM2.5 for 24 h. (A) The NO concentration in the culture medium was determined by the Griess reaction. (B-D) The PGE2 (B), IL-6 (C), and IL-1β (D) concentration was determined using commercial ELISA kits. The absorbance was measured using a microplate reader. The error bars represent the mean ± SD of 3 independent experiments.SF, Schisandrae Fructus ethanol extract; PM, particulate matter; NO, nitric oxide; PGE2, prostaglandin E2; IL, interleukin; ELISA, enzyme-linked immunosorbent assay.
***P < 0.001 vs. PM2.5-unstimulated cells; #P < 0.05, ##P < 0.01 and ###P < 0.001 vs. PM2.5-stimulated cells.
|
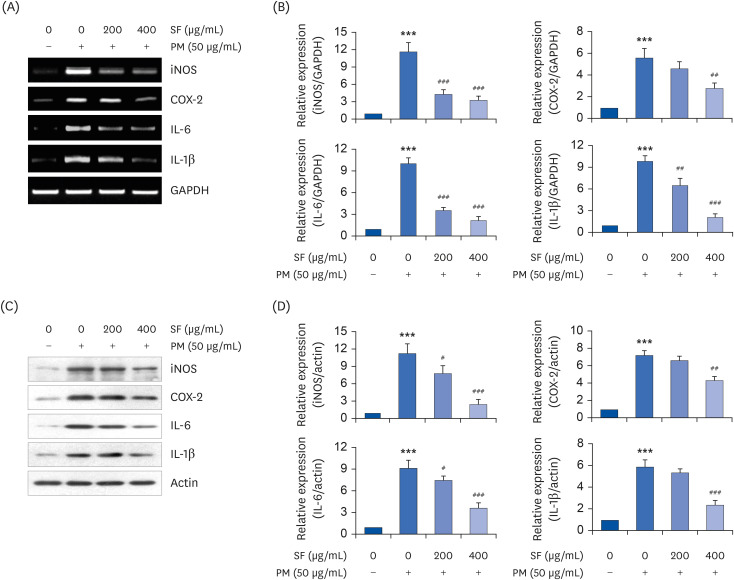 | Fig. 2Effect of SF on the expression of pro-inflammatory enzymes and cytokines in PM2.5-stimulated RAW 264.7 macrophages. Cells were treated with the indicated concentrations of SF for 1 h and then stimulated with 50 µg/mL PM2.5 for 24 h. After treatment, total RNA and protein were extracted from the cells. The expression levels of iNOS, COX-2, IL-6, and IL-1β mRNA (A) and proteins (C) were measured by RT-PCR and Western blot analysis, respectively. GAPDH and actin and were used as internal controls for the RT-PCR and Western blot analyses, respectively. (B, D) Bands were quantified using ImageJ and normalized to GAPDH and actin, and the ratio was determined. Data are expressed as the mean ± SD of 3 independent experiments.SF, Schisandrae Fructus ethanol extract; PM, particulate matter; iNOS, inducible nitric oxide synthase; COX-2, cyclooxygenase-2; IL, interleukin; RT-PCR, reverse transcription-polymerase chain reaction; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
***P < 0.001 vs. PM-unstimulated cells; #P < 0.05, ##P < 0.01 and ###P < 0.001 vs. PM-stimulated cells.
|
SF reduces the production and expression of PM2.5-induced pro-inflammatory cytokines in RAW 264.7 macrophages
SF suppresses the nuclear translocation of NF-κB in PM2.5-stimulated RAW 264.7 macrophages
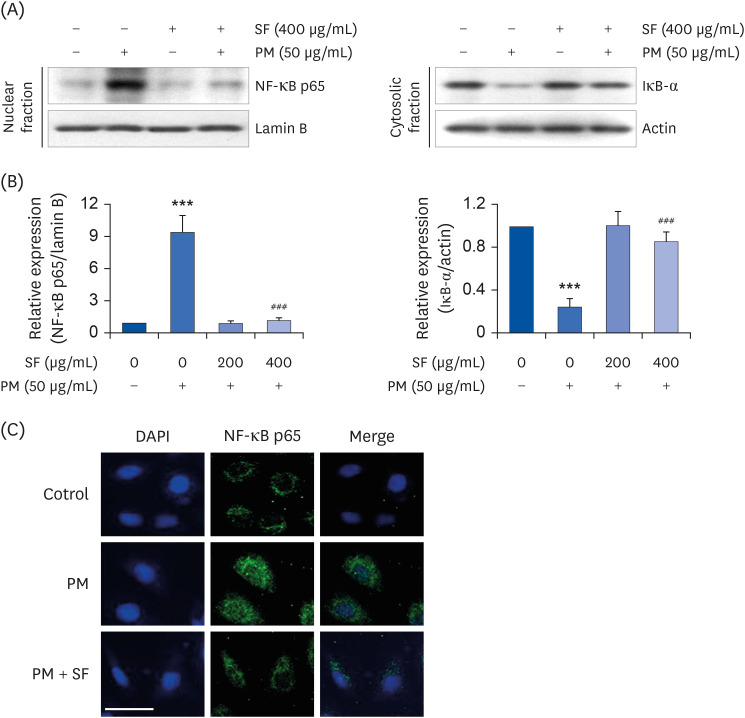 | Fig. 3Inactivation of NF-κB signaling pathway by SF in PM2.5-stimulated RAW 264.7 macrophages. Cells were treated with 400 µg/mL SF alone for 24 h or pre-treated with or without 400 µg/mL SF for 1 h before 50 µg/mL PM2.5 stimulation for 1 h. (A) For Western blot analysis, nuclear and cytosolic proteins were isolated, and the expression of NF-κB and IκB-a was investigated. Protein loading was confirmed by the analysis of lamin B or actin expression in each protein extract.(B) Bands were quantified using ImageJ and normalized to lamin B and actin, and the ratio was determined. Data are expressed as the mean ± SD of 3 independent experiments. (C) The cells were subjected to immunofluorescence staining with NF-κB p65 antibody and representative fluorescence images were acquired using a fluorescence microscope. Green fluorescence indicates the localization of NF-κB p65 and blue fluorescence by DAPI staining allows visualization of the nuclei (scale bar = 200 µM).SF, Schisandrae Fructus ethanol extract; PM, particulate matter; NF-κB, nuclear factor-kappa B; DAPI, 4′,6-diamidino-2-phenylindole.
***P < 0.001 vs. PM-unstimulated cells; ###P < 0.001 vs. PM-stimulated cells.
|
SF alleviates the PM2.5-mediated generation of ROS in RAW 264.7 macrophages
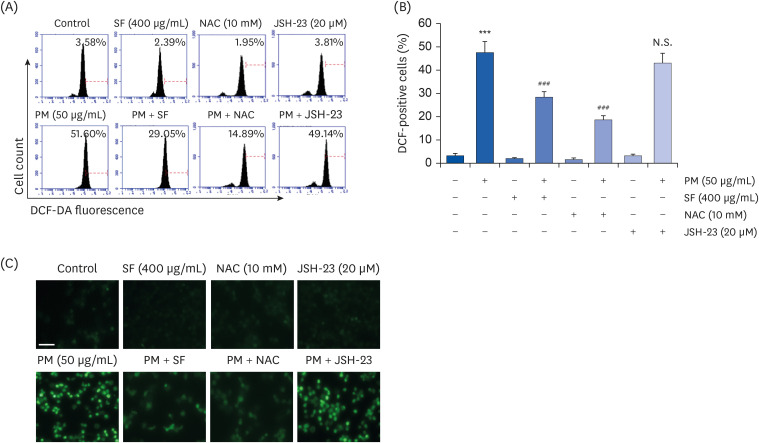 | Fig. 4Inhibition of ROS generation by SF in PM2.5-stimulated RAW 264.7 macrophages. Cells were pre-treated with 400 µg/mL SF, 10 mM NAC or 20 µM JSH-23 for 1 h and then treated with 50 µg/mL PM2.5 for 1 h. (A) The DCF-DA-stained cells were collected, and then DCF fluorescence was analyzed by flow cytometry. (B) Data are given as the mean ± SD of 3 independent experiments. (C) ROS generation was also detected by a fluorescence microscope and representative fluorescence micrographs depicting ROS generation are presented. Green fluorescence indicates the intensity of ROS generation (scale bar = 200 µM).ROS, reactive oxygen species; SF, Schisandrae Fructus ethanol extract; NAC, N-acetyl cysteine; JSH-23, 4-methyl-N
1-(3-phenyl-propyl)-benzene-1,2-diamine; DCF-DA, 5,6-carboxy-2′,7′-dichlorofluorescein diacetate; N.S., not significant; PM, particulate matter.
***P < 0.001 vs. PM2.5-unstimulated cells; ###P < 0.001 vs. PM2.5-stimulated cells.
|
The inhibitory effect of SF on PM2.5-induced NF-κB activation and inflammatory response is ROS-dependent in RAW 264.7 macrophages
 | Fig. 5Role of ROS on the inhibitory effect of SF on PM2.5-induced NF-κB activation and inflammatory response. Cells were pre-treated with 400 µg/mL SF or 10 mM NAC for 1 h and then treated with 50 µg/mL PM2.5 for 1 h (A, B) or 24 h (C, D). (A, B) Nuclear and cytosolic proteins were isolated, and the expression of NF-κB and IκB-a was investigated. Protein loading was confirmed by the analysis of lamin B or actin expression in each protein extract. (B) Bands were quantified using ImageJ and normalized to lamin B and actin, and the ratio was determined. The NO (C) and IL-6 (D) concentration in the culture medium was determined by the Griess reaction and IL-6 ELISA kit, respectively. The absorbance was measured using a microplate reader. (B-D) Data are expressed as the mean ± SD of 3 independent experiments.ROS, reactive oxygen species; SF, Schisandrae Fructus ethanol extract; PM, particulate matter; NF-κB, nuclear factor-kappa B; NO, nitric oxide; IL, interleukin; ELISA, enzyme-linked immunosorbent assay.
***P < 0.001 vs. PM-unstimulated cells; ##P < 0.01 and ###P < 0.001 vs. PM-stimulated cells.
|
SF weakens the production of NO and ROS in PM2.5-treated zebrafish larvae
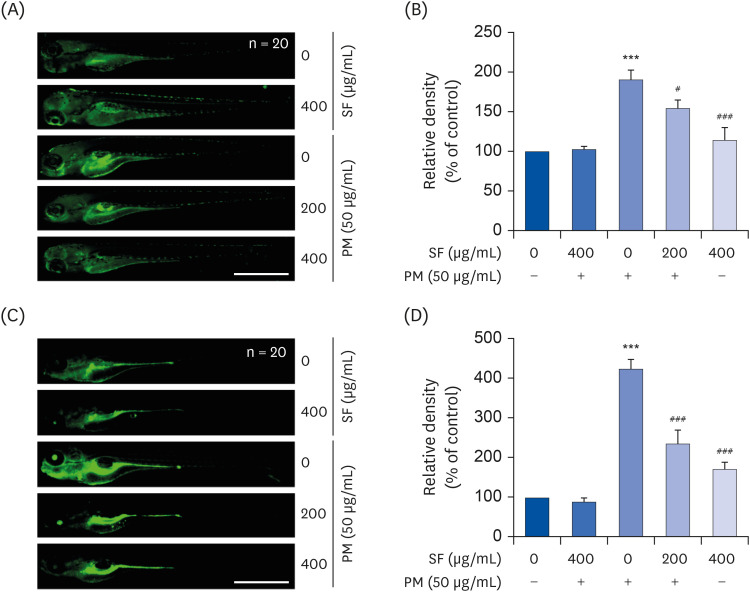 | Fig. 6Inhibition of PM2.5-induced NO and ROS generation by SF in zebrafish larvae. Zebrafish at 3 dpf were treated with 50 µg/mL PM2.5 and placed in E3 media containing the indicated concentrations of SF for 24 h. The larvae were incubated with 5 µM DAF-FM-DA (A, B) or 20 µM DCF-DA (C, D) for NO and ROS detection and visualized using the CELENA® S Digital Imaging System (scale bar = 1,000 µm). (B, D) Relative fluorescence intensities were calculated and expressed compared to the untreated control. Each value indicates the mean ± SD of 3 independent experiments. Significant differences among the groups were determined.PM, particulate matter; NO, nitric oxide; ROS, reactive oxygen species; SF, Schisandrae Fructus ethanol extract; dpf, days post-fertilized; DAF-FM-DA, 4-amino-5-methylamino-2′7′-difluorofluorescein diacetate; DCF-DA, 5,6-carboxy-2′,7′-dichlorofluorescein diacetate.
***P < 0.001 vs. PM2.5-unstimulated larvae; #P < 0.05 and ###P < 0.001 vs. PM2.5-stimulated larvae.
|




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download