Abstract
Food allergy is a growing global public health concern. As treatment strategies are currently limited to allergen avoidance and emergency interventions, there is an increasing demand for appropriate models of food allergy for the development of new therapeutics. Many models of food allergy rely heavily on the use of animals, and while useful, many are unable to accurately reflect the human system. In order to bridge the gap between in vivo animal models and clinical trials with human patients, human models of food allergy are of great importance. This review will summarize the commonly used human ex vivo and in vitro models of food allergy and highlight their advantages and limitations regarding how accurately they represent the human in vivo system. We will cover biopsy-based systems, precision cut organ slices, and coculture systems as well as organoids and organ-on-a-chip. The availability of appropriate experimental models will allow us to move forward in the field of food allergy research, to search for effective treatment options and to further explore the cause and progression of this disorder.
Go to : 
References
1. Savage J, Johns CB. Food allergy: epidemiology and natural history. Immunol Allergy Clin North Am. 2015; 35:45–59.
2. Yu W, Freeland DMH, Nadeau KC. Food allergy: immune mechanisms, diagnosis and immunotherapy. Nat Rev Immunol. 2016; 16:751–65.

3. Prescott SL, Pawankar R, Allen KJ, Campbell DE, Sinn JKh, Fiocchi A, Ebisawa M, Sampson HA, Beyer K, Lee BW. A global survey of changing patterns of food allergy burden in children. World Allergy Organ J. 2013; 6:21.

4. Benedé S, Blázquez AB, Chiang D, Tordesillas L, Berin MC. The rise of food allergy: environmental factors and emerging treatments. EBioMedicine. 2016; 7:27–34.

5. Yang MS, Kim JY, Kim BK, Park HW, Cho SH, Min KU, Kang HR. True rise in anaphylaxis incidence: Epidemiologic study based on a national health insurance database. Medicine (Baltimore). 2017; 96:e5750.
6. Valenta R, Hochwallner H, Linhart B, Pahr S. Food allergies: the basics. Gastroenterology. 2015; 148:1120–31. e4.

7. Hong X, Hao K, Ladd-Acosta C, Hansen KD, Tsai HJ, Liu X, Xu X, Thornton TA, Caruso D, Keet CA, Sun Y, Wang G, Luo W, Kumar R, Fuleihan R, Singh AM, Kim JS, Story RE, Gupta RS, Gao P, Chen Z, Walker SO, Bartell TR, Beaty TH, Fallin MD, Schleimer R, Holt PG, Nadeau KC, Wood RA, Pongracic JA, Weeks DE, Wang X. Genome-wide association study identifies peanut allergy-specific loci and evidence of epigenetic mediation in US children. Nat Commun. 2015; 6:6304.

8. Portelli MA, Hodge E, Sayers I. Genetic risk factors for the development of allergic disease identified by genome-wide association. Clin Exp Allergy. 2015; 45:21–31.

9. Barcik W, Untersmayr E, Pali-Schöll I, O'Mahony L, Frei R. Influence of microbiome and diet on immune responses in food allergy models. Drug Discov Today Dis Models. 2015; 17-18:71–80.

11. Conrad DH, Ford JW, Sturgill JL, Gibb DR. CD23: an overlooked regulator of allergic disease. Curr Allergy Asthma Rep. 2007; 7:331–7.

12. Hussain M, Epstein MM, Noti M. Experimental food allergy models to study the role of innate immune cells as initiators of allergen-specific Th2 immune responses. Drug Discov Today Dis Model. 2016; 17-18:55–62.

13. Fujiwara S. Humanized mice: A brief overview on their diverse applications in biomedical research. J Cell Physiol. 2018; 233:2889–901.

14. Shin JS, Greer AM. The role of Fcε RI expressed in dendritic cells and monocytes. Cell Mol Life Sci. 2015; 72:2349–60.

15. Platzer B, Baker K, Vera MP, Singer K, Panduro M, Lexmond WS, Turner D, Vargas SO, Kinet JP, Maurer D, Baron RM, Blumberg RS, Fiebiger E. Dendritic cell-bound IgE functions to restrain allergic inflammation at mucosal sites. Mucosal Immunol. 2015; 8:516–32.

16. Maurer D, Fiebiger S, Ebner C, Reininger B, Fischer GF, Wichlas S, Jouvin MH, Schmitt-Egenolf M, Kraft D, Kinet JP, Stingl G. Peripheral blood dendritic cells express Fc epsilon RI as a complex composed of Fc epsilon RI alpha- and Fc epsilon RI gamma-chains and can use this receptor for IgE-mediated allergen presentation. J Immunol. 1996; 157:607–16.
17. Blume C, Davies DE. In vitro and ex vivo models of human asthma. Eur J Pharm Biopharm. 2013; 84:394–400.

18. Wang J, Jones SM, Pongracic JA, Song Y, Yang N, Sicherer SH, Makhija MM, Robison RG, Moshier E, Godbold J, Sampson HA, Li XM. Safety, clinical, and immunologic efficacy of a Chinese herbal medicine (Food Allergy Herbal Formula-2) for food allergy. J Allergy Clin Immunol. 2015; 136:962–70. e1.

19. Wood RA, Sicherer SH, Burks AW, Grishin A, Henning AK, Lindblad R, Stablein D, Sampson HA. A phase 1 study of heat/phenol-killed, E. coli-encapsulated, recombinant modified peanut proteins Ara h 1, Ara h 2, and Ara h 3 (EMP-123) for the treatment of peanut allergy. Allergy. 2013; 68:803–8.
20. Bryce PJ, Falahati R, Kenney LL, Leung J, Bebbington C, Tomasevic N, Krier RA, Hsu CL, Shultz LD, Greiner DL, Brehm MA. Humanized mouse model of mast cell-mediated passive cutaneous anaphylaxis and passive systemic anaphylaxis. J Allergy Clin Immunol. 2016; 138:769–79.

21. Pagovich OE, Wang B, Chiuchiolo MJ, Kaminsky SM, Sondhi D, Jose CL, Price CC, Brooks SF, Mezey JG, Crystal RG. Anti-hIgE gene therapy of peanut-induced anaphylaxis in a humanized murine model of peanut allergy. J Allergy Clin Immunol. 2016; 138:1652–62. e7.

22. Burton OT, Stranks AJ, Tamayo JM, Koleoglou KJ, Schwartz LB, Oettgen HC. A humanized mouse model of anaphylactic peanut allergy. J Allergy Clin Immunol. 2017; 139:314–22. e9.

23. Jaffar ZH, Stanciu L, Pandit A, Lordan J, Holgate ST, Roberts K. Essential role for both CD80 and CD86 costimulation, but not CD40 interactions, in allergen-induced Th2 cytokine production from asthmatic bronchial tissue: role for alphabeta, but not gammadelta, T cells. J Immunol. 1999; 163:6283–91.
24. Hidi R, Riches V, Al-Ali M, Cruikshank WW, Center DM, Holgate ST, Djukanovic R. Role of B7-CD28/CTLA-4 costimulation and NF-kappa B in allergen-induced T cell chemotaxis by IL-16 and RANTES. J Immunol. 2000; 164:412–8.
25. Lordan JL, Davies DE, Wilson SJ, Dent G, Corkhill A, Jaffar Z, Roberts K, Djukanović R, Holgate ST. The role of CD28-B7 costimulation in allergen-induced cytokine release by bronchial mucosa from patients with moderately severe asthma. J Allergy Clin Immunol. 2001; 108:976–81.

26. Vijayanand P, Durkin K, Hartmann G, Morjaria J, Seumois G, Staples KJ, Hall D, Bessant C, Bartholomew M, Howarth PH, Friedmann PS, Djukanovic R. Chemokine receptor 4 plays a key role in T cell recruitment into the airways of asthmatic patients. J Immunol. 2010; 184:4568–74.

27. Jaffar Z, Roberts K, Pandit A, Linsley P, Djukanovic R, Holgate S. B7 costimulation is required for IL-5 and IL-13 secretion by bronchial biopsy tissue of atopic asthmatic subjects in response to allergen stimulation. Am J Respir Cell Mol Biol. 1999; 20:153–62.

28. Pizzuti D, Senzolo M, Buda A, Chiarelli S, Giacomelli L, Mazzon E, Curioni A, Faggian D, De Lazzari F. In vitro model for IgE mediated food allergy. Scand J Gastroenterol. 2011; 46:177–87.
29. Reimann HJ, Lewin J. Gastric mucosal reactions in patients with food allergy. Am J Gastroenterol. 1988; 83:1212–9.
30. Reimann HJ, Ring J, Ultsch B, Wendt P. Intragastral provocation under endoscopic control (IPEC) in food allergy: mast cell and histamine changes in gastric mucosa. Clin Allergy. 1985; 15:195–202.

31. Bischoff SC, Mayer J, Wedemeyer J, Meier PN, Zeck-Kapp G, Wedi B, Kapp A, Cetin Y, Gebel M, Manns MP. Colonoscopic allergen provocation (COLAP): a new diagnostic approach for gastrointestinal food allergy. Gut. 1997; 40:745–53.

32. Bach PH, Vickers AE, Fisher R, Baumann A, Brittebo E, Carlile DJ, Koster HJ, Lake BG, Salmon F, Sawyer TW, Skabinsky G. The use of tissue slices in pharmacotoxicology studies. The report and recommendations of ECVAM workshop 20. Altern Lab Anim. 1996; 24:893–923.
33. de Kanter R, Tuin A, van de Kerkhof E, Martignoni M, Draaisma AL, de Jager MH, de Graaf IA, Meijer DK, Groothuis GM. A new technique for preparing precision-cut slices from small intestine and colon for drug biotransformation studies. J Pharmacol Toxicol Methods. 2005; 51:65–72.

34. Sewald K, Braun A. Assessment of immunotoxicity using precision-cut tissue slices. Xenobiotica. 2013; 43:84–97.

35. Wohlsen A, Martin C, Vollmer E, Branscheid D, Magnussen H, Becker WM, Lepp U, Uhlig S. The early allergic response in small airways of human precision-cut lung slices. Eur Respir J. 2003; 21:1024–32.

36. Sanderson MJ. Exploring lung physiology in health and disease with lung slices. Pulm Pharmacol Ther. 2011; 24:452–65.

37. Henjakovic M, Sewald K, Switalla S, Kaiser D, Müller M, Veres TZ, Martin C, Uhlig S, Krug N, Braun A. Ex vivo testing of immune responses in precision-cut lung slices. Toxicol Appl Pharmacol. 2008; 231:68–76.

38. Lamb DJ, Wollin SL, Schnapp A, Bischoff D, Erb KJ, Bouyssou T, Guilliard B, Strasser C, Wex E, Blum S, Thaler E, Nickel H, Radmacher O, Haas H, Swantek JL, Souza D, Canfield M, White D, Panzenbeck M, Kashem MA, Sanville-Ross M, Kono T, Sewald K, Braun A, Obernolte H, Danov O, Schaenzle G, Rast G, Maier GM, Hoffmann M. BI 1002494, a novel potent and selective oral spleen tyrosine kinase inhibitor, displays differential potency in human basophils and B cells. J Pharmacol Exp Ther. 2016; 357:554–61.

39. Groothuis GM, de Graaf IA. Precision-cut intestinal slices as in vitro tool for studies on drug metabolism. Curr Drug Metab. 2013; 14:112–9.
40. Graaf IA, Groothuis GM, Olinga P. Precision-cut tissue slices as a tool to predict metabolism of novel drugs. Expert Opin Drug Metab Toxicol. 2007; 3:879–98.

41. Pham BT, van Haaften WT, Oosterhuis D, Nieken J, de Graaf IA, Olinga P. Precision-cut rat, mouse, and human intestinal slices as novel models for the early-onset of intestinal fibrosis. Physiol Rep. 2015 Apr; 3(4):). pii: e12323.https://doi.org/10.14814/phy2.12323.

42. Li M, de Graaf IA, Groothuis GM. Precision-cut intestinal slices: alternative model for drug transport, metabolism, and toxicology research. Expert Opin Drug Metab Toxicol. 2016; 12:175–90.

43. de Graaf IA, Olinga P, de Jager MH, Merema MT, de Kanter R, van de Kerkhof EG, Groothuis GM. Preparation and incubation of precision-cut liver and intestinal slices for application in drug metabolism and toxicity studies. Nat Protoc. 2010; 5:1540–51.

44. Pichavant M, Charbonnier AS, Taront S, Brichet A, Wallaert B, Pestel J, Tonnel AB, Gosset P. Asthmatic bronchial epithelium activated by the proteolytic allergen Der p 1 increases selective dendritic cell recruitment. J Allergy Clin Immunol. 2005; 115:771–8.

45. Yamashita S, Yokoyama Y, Hashimoto T, Mizuno M. A novel in vitro co-culture model comprised of Caco-2/RBL-2H3 cells to evaluate anti-allergic effects of food factors through the intestine. J Immunol Methods. 2016; 435:1–6.

46. de Kivit S, Tobin MC, DeMeo MT, Fox S, Garssen J, Forsyth CB, Keshavarzian A, Landay AL. In vitro evaluation of intestinal epithelial TLR activation in preventing food allergic responses. Clin Immunol. 2014; 154:91–9.

47. Barkauskas CE, Chung MI, Fioret B, Gao X, Katsura H, Hogan BL. Lung organoids: current uses and future promise. Development. 2017; 144:986–97.

48. Merker SR, Weitz J, Stange DE. Gastrointestinal organoids: how they gut it out. Dev Biol. 2016; 420:239–50.

49. Uchida H, Machida M, Miura T, Kawasaki T, Okazaki T, Sasaki K, Sakamoto S, Ohuchi N, Kasahara M, Umezawa A, Akutsu H. A xenogeneic-free system generating functional human gut organoids from pluripotent stem cells. JCI Insight. 2017; 2:e86492.

50. Hill DR, Spence JR. Gastrointestinal organoids: understanding the molecular basis of the host-microbe interface. Cell Mol Gastroenterol Hepatol. 2016; 3:138–49.

51. Nichols JE, Niles JA, Vega SP, Cortiella J. Novel in vitro respiratory models to study lung development, physiology, pathology and toxicology. Stem Cell Res Ther. 2013; 4(Suppl 1):S7.

52. Nesmith AP, Agarwal A, McCain ML, Parker KK. Human airway musculature on a chip: an in vitro model of allergic asthmatic bronchoconstriction and bronchodilation. Lab Chip. 2014; 14:3925–36.

53. Kim HJ, Ingber DE. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr Biol (Camb). 2013; 5:1130–40.

54. Kim HJ, Huh D, Hamilton G, Ingber DE. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012; 12:2165–74.

55. Kim HJ, Li H, Collins JJ, Ingber DE. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci U S A. 2016; 113:E7–15.

Go to : 
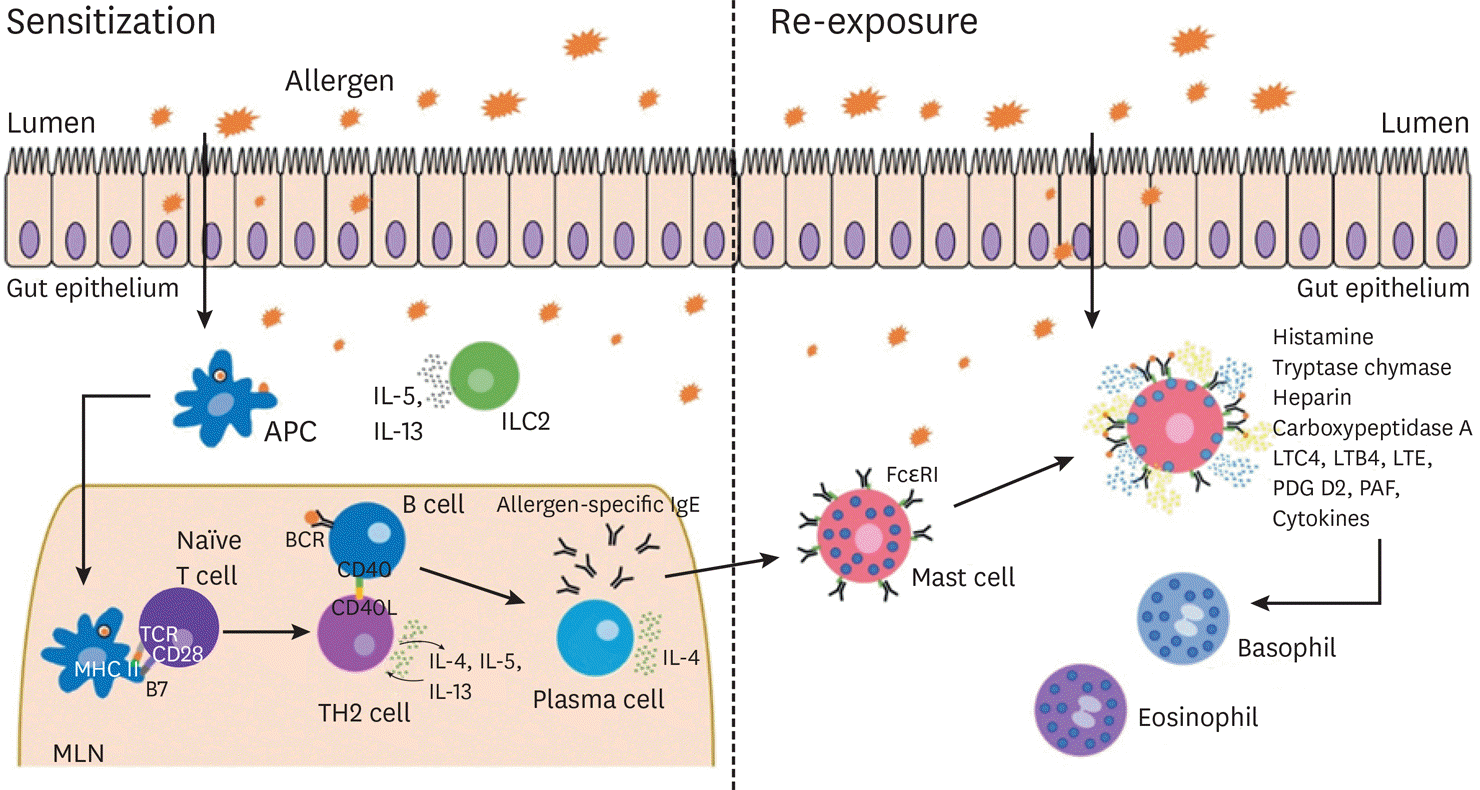 | Fig. 1.Schematic of allergic sensitization and re-exposure. Sensitization: Food allergens can cross the luminal barrier by transcellular or paracellular transport, or through direct luminal antigen sampling by dendritic cells (DCs). These allergens are picked up and processed by DCs which can then migrate to mesenteric lymph nodes (MLN) and present the allergen to naïve CD4+ T cells in the context of MHC II. These T cells can then differentiate into Th2 cells that secrete pro-allergic cytokines that influence B cells to become IgE secreting plasma cells. These allergen-specific IgE antibodies can then bind to the Fcε RI (high affinity IgE receptor) expressed on the main effector cells of the allergic response, including mast cells and basophils. Re-sensitization: Upon re-exposure to the allergen, the allergen-specific IgE on the surface of the effector cells can bind, crosslink and activate the cell leading to degranulation of pro-inflammatory mediators. The release of these factors can cause multiple downstream effects and result in the recruitment of other inflammatory cell subsets. APC, antigen-presenting cell; IL, interleukin; MHC, major histocompatibility complex; LTC4, leukotriene C4; LTB4, leukotriene B4; LTE, leukotriene E; PDG D2, prostaglandin D2; PAF, platelet-activating factor. |
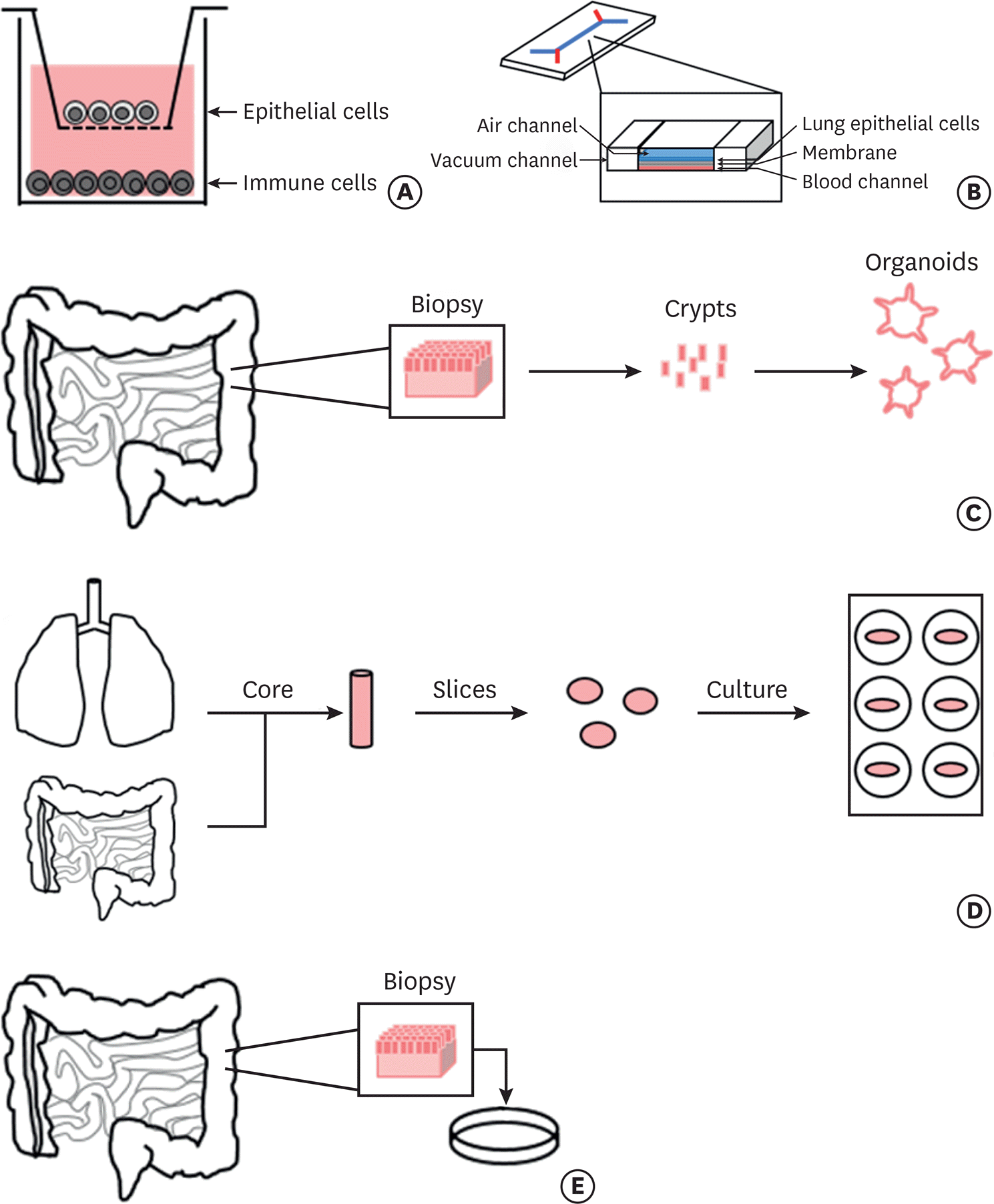 | Fig. 2.Human in vitro and ex vivo models for food allergy research. Different types of human tissue and cell-based models potentially used to study food allergy in order of proximity to the human system. (A) In vitro coculture systems utilize Transwell inserts on which epithelial cells are cultured and allowed to polarize. Immune cells can be preloaded into the wells to examine cellular interactions. (B) Lung-on-a chip models consist of a membrane on which human lung epithelial cells line the top and are separated from human blood cells that line the bottom. Air flows over the epithelial cells and fluid that mimics blood flows along the bottom creating an environment reflective of the human lung. Vacuum channels on either side allow the cells to be stretched gently mimicking the mechanical movements of breathing. (C) Organoids can be generated using gut samples from human donors, the crypts are then isolated and used to form mini-gut cultures [57]. (D) Precision cut organ slices require the formation of tissue cores from agarose filled organs that are then cut into thin slices of fixed thickness. These slices are then incubated and used for experimentation. (E) Biopsy based models utilize tissue directly extracted from the organ of interest which is subsequently cultured and used for experimentation. |
Table 1.
Comparison of the different human in vitro and ex vivo models of food allergy




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download