Abstract
The pandemic of avian influenza viruses (AIVs) in Asia has caused enormous economic loss in poultry industry and human health threat, especially clade 2.3.4.4 H5 and H7 subtypes in recent years. The endemic chicken H6 virus in Taiwan has also brought about human and dog infections. Since wild waterfowls is the major AIV reservoir, it is important to monitor the diversified subtypes in wildfowl flocks in early stage to prevent viral reassortment and transmission. To develop a more efficient and sensitive approach is a key issue in epidemic control. In this study, we integrate multiplex reverse transcription recombinase polymerase amplification (RT-RPA) and capillary electrophoresis (CE) for high-throughput detection and differentiation of AIVs in wild waterfowls in Taiwan. Four viral genes were detected simultaneously, including nucleoprotein (NP) gene of all AIVs, hemagglutinin (HA) gene of clade 2.3.4.4 H5, H6 and H7 subtypes. The detection limit of the developed detection system could achieve as low as one copy number for each of the four viral gene targets. Sixty wild waterfowl field samples were tested and all of the four gene signals were unambiguously identified within 6 h, including the initial sample processing and the final CE data analysis. The results indicated that multiplex RT-RPA combined with CE was an excellent alternative for instant simultaneous AIV detection and subtype differentiation. The high efficiency and sensitivity of the proposed method could greatly assist in wild bird monitoring and epidemic control of poultry.
Go to : 
References
1. Lamb RA, Krug RM. Orthomyxoviridae: the viruses and their replication. Fields BN, Knipe DM, Howley PM, Chanock RM, Melnick JL, Momath TP, Roizman B, editors. (eds.).Fundamental Virology. p. 605–648. Lippincott-Raven Publisher;Philadelphia: 1996.
2. Webster RG, Shortridge KF, Kawaoka Y. Influenza: interspecies transmission and emergence of new pandemics. FEMS Immunol Med Microbiol. 1997; 18:275–279.

3. Chang CF, King CC, Wan CH, Chang YC, Chan TC, David Lee CC, Chou PH, Li ZR, Li YT, Tseng TJ, Lee PF, Chang CH. Lessons from the largest epidemic of avian influenza viruses in Taiwan, 2015. Avian Dis. 2016; 60(Suppl):156–171.

4. Lee MS, Chen LH, Chen YP, Liu YP, Li WC, Lin YL, Lee F. Highly pathogenic avian influenza viruses H5N2, H5N3, and H5N8 in Taiwan in 2015. Vet Microbiol. 2016; 187:50–57.

5. Bureau of Animal and Plant Health Inspection and Quarantine. [cited 2015 August 8]. Available from:. http://ai.gov.tw.
6. Lee MS, Chang PC, Shien JH, Cheng MC, Chen CL, Shieh HK. Genetic and pathogenic characterization of H6N1 avian influenza viruses isolated in Taiwan between 1972 and 2005. Avian Dis. 2006; 50:561–571.

7. Wei SH, Yang JR, Wu HS, Chang MC, Lin JS, Lin CY, Liu YL, Lo YC, Yang CH, Chuang JH, Lin MC, Chung WC, Liao CH, Lee MS, Huang WT, Chen PJ, Liu MT, Chang FY. Human infection with avian influenza A H6N1 virus: an epidemiological analysis. Lancet Respir Med. 2013; 1:771–778.

8. Lin HT, Wang CH, Chueh LL, Su BL, Wang LC. Influenza A(H6N1) virus in dogs, Taiwan. Emerg Infect Dis. 2015; 21:2154–2157.

9. Zhu H, Lam TT, Smith DK, Guan Y. Emergence and development of H7N9 influenza viruses in China. Curr Opin Virol. 2016; 16:106–113.

10. Lee DH, Bertran K, Kwon JH, Swayne DE. Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3.4.4. J Vet Sci. 2017; 18(S1):269–280.

11. Daher RK, Stewart G, Boissinot M, Bergeron MG. Recombinase polymerase amplification for diagnostic applications. Clin Chem. 2016; 62:947–958.

12. Jaroenram W, Owens L. Recombinase polymerase amplification combined with a lateral flow dipstick for discriminating between infectious Penaeus stylirostris densovirus and virus-related sequences in shrimp genome. J Virol Methods. 2014; 208:144–151.

13. Piepenburg O, Williams CH, Stemple DL, Armes NA. DNA detection using recombination proteins. PLoS Biol. 2006; 4:e204.

14. Euler M, Wang Y, Nentwich O, Piepenburg O, Hufert FT, Weidmann M. Recombinase polymerase amplification assay for rapid detection of Rift Valley fever virus. J Clin Virol. 2012; 54:308–312.

15. Abd El Wahed A, Weidmann M, Hufert FT. Diagnostics-in-a-suitcase: development of a portable and rapid assay for the detection of the emerging avian influenza A (H7N9) virus. J Clin Virol. 2015; 69:16–21.

16. Yehia N, Arafa AS, Abd El Wahed A, El-Sanousi AA, Weidmann M, Shalaby MA. Development of reverse transcription recombinase polymerase amplification assay for avian influenza H5N1 HA gene detection. J Virol Methods. 2015; 223:45–49.

17. Daher RK, Stewart G, Boissinot M, Boudreau DK, Bergeron MG. Influence of sequence mismatches on the specificity of recombinase polymerase amplification technology. Mol Cell Probes. 2015; 29:116–121.

18. McMurray CL, Hardy KJ, Hawkey PM. Rapid, automated epidemiological typing of methicillin-resistant Staphylococcus aureus. J Microbiol Methods. 2010; 80:109–111.
19. Jiang LX, Ren HY, Zhou HJ, Zhao SH, Hou BY, Yan JP, Qin T, Chen Y. Simultaneous detection of 13 key bacterial respiratory pathogens by combination of multiplex PCR and capillary electrophoresis. Biomed Environ Sci. 2017; 30:549–561.
20. Wu XL, Xiao L, Lin H, Yang M, Chen SJ, An W, Wang Y, Yao XP, Yang ZX, Tang ZZ. A novel capillary electrophoresis-based high-throughput multiplex polymerase chain reaction system for the simultaneous detection of nine pathogens in swine. BioMed Res Int. 2017; 2017:7243909.

21. Schroeder ME, Johnson DJ, Ostlund EN, Meier J, Bounpheng MA, Clavijo A. Development and performance evaluation of a streamlined method for nucleic acid purification, denaturation, and multiplex detection of Bluetongue virus and Epizootic hemorrhagic disease virus. J Vet Diagn Invest. 2013; 25:709–719.

22. Nikolayevskyy V, Trovato A, Broda A, Borroni E, Cirillo D, Drobniewski F. MIRU-VNTR genotyping of mycobacterium tuberculosis strains using QIAxcel technology: a multicentre evaluation study. PLoS One. 2016; 11:e0149435.

23. Dean DA, Wadl PA, Hadziabdic D, Wang X, Trigiano RN. Analyzing microsatellites using the QIAxcel system. Methods Mol Biol. 2013; 1006:223–243.

24. Barakat H, El-Garhy HA, Moustafa MM. Detection of pork adulteration in processed meat by species-specific PCR-QIAxcel procedure based on D-loop and cytb genes. Appl Microbiol Biotechnol. 2014; 98:9805–9816.

25. Kerékgyártó M, Kerekes T, Tsai E, Amirkhanian VD, Guttman A. Light-emitting diode induced fluorescence (LED-IF) detection design for a pen-shaped cartridge based single capillary electrophoresis system. Electrophoresis. 2012; 33:2752–2758.

26. Food and Agriculture Organization of the United Nations. Wild Birds and Avian Influenza: an Introduction to Applied Field Research and Disease Sampling Techniques. FAO;Rome: 2007.
27. Fereidouni SR, Harder TC, Gaidet N, Ziller M, Hoffmann B, Hammoumi S, Globig A, Starick E. Saving resources: avian influenza surveillance using pooled swab samples and reduced reaction volumes in real-time RT-PCR. J Virol Methods. 2012; 186:119–125.

28. Thermo Fisher Scientific. Multiple Primer Analyzer [Internet]. Thermo Fisher Scientific;2015. [updated 2015;cited 2015 August 8]. Available from:. https://www.thermofisher.com/tw/zt/home/brands/thermo-scientific/molecular-biology/molecular-biology-learning-center/molecular-biology-resource-library/thermo-scientific-web-tools/multiple-primer-analyzer.html.
29. Yang Y, Qin X, Song Y, Zhang W, Hu G, Dou Y, Li Y, Zhang Z. Development of real-time and lateral flow strip reverse transcription recombinase polymerase amplification assays for rapid detection of peste des petits ruminants virus. Virol J. 2017; 14:24.

30. Crannell Z, Castellanos-Gonzalez A, Nair G, Mejia R, White AC, Richards-Kortum R. Multiplexed recombinase polymerase amplification assay to detect intestinal protozoa. Anal Chem. 2016; 88:1610–1616.

31. Kersting S, Rausch V, Bier FF, von Nickisch-Rosenegk M. Multiplex isothermal solid-phase recombinase polymerase amplification for the specific and fast DNA-based detection of three bacterial pathogens. Mikrochim Acta. 2014; 181:1715–1723.

32. Luo GC, Yi TT, Jiang B, Guo XL, Zhang GY. Betaine-assisted recombinase polymerase assay with enhanced specificity. Anal Biochem. 2019; 575:36–39.

33. Steel J, Lowen AC. Influenza A virus reassortment. Compans RW, Oldstone MBA, editors. (eds.).Influenza Pathogenesis and Control-Volume I. pp.p. 377–401. Springer;Heidelberg: 2014.
Go to : 
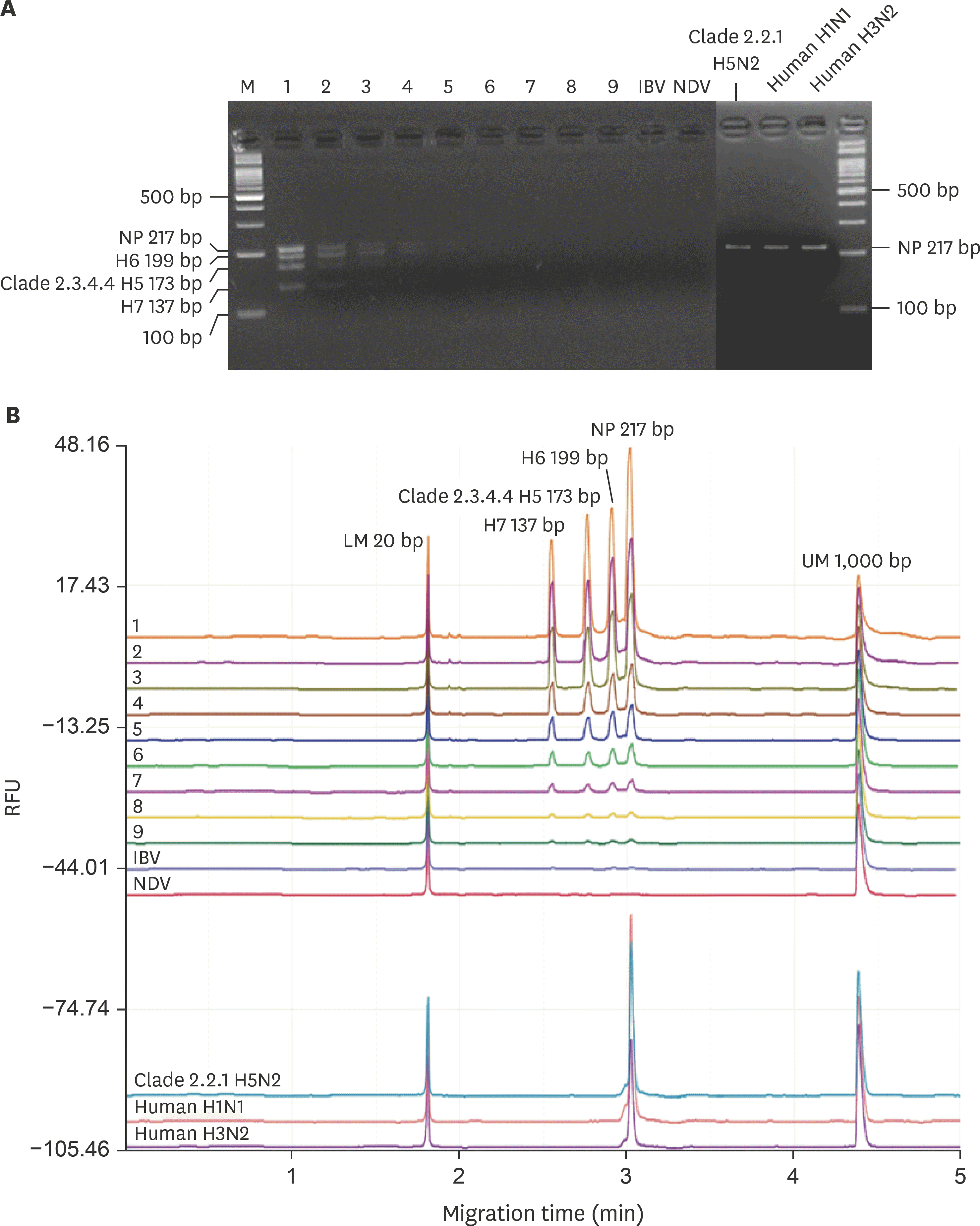 | Fig. 1.Detection limit comparison between traditional plate agarose gel electrophoresis (A) and capillary electrophoresis (B). In vitro transcribed RNA from each targeted viral gene was quantified and used for multiplex reverse transcription recombinase polymerase amplification reaction templates. M, 100 bp ladder marker; LM, lower alignment marker (size = 20 bp); UM, upper alignment marker (size = 1,000 bp); 1, 108 copy numbers; 2, 107 copy numbers; 3, 106 copy numbers; 4, 105 copy numbers; 5, 104 copy numbers; 6, 103 copy numbers; 7, 102 copy numbers; 8, 101 copy numbers; 9, 100 copy numbers; IBV, infectious bronchitis virus; NDV, Newcastle disease virus. Other common avian respiratory viruses (IBV and NDV) and type A influenza viruses (clade 2.2.1 low pathogenic H5N2, human H1N1 and human H3N2) were used as control. |
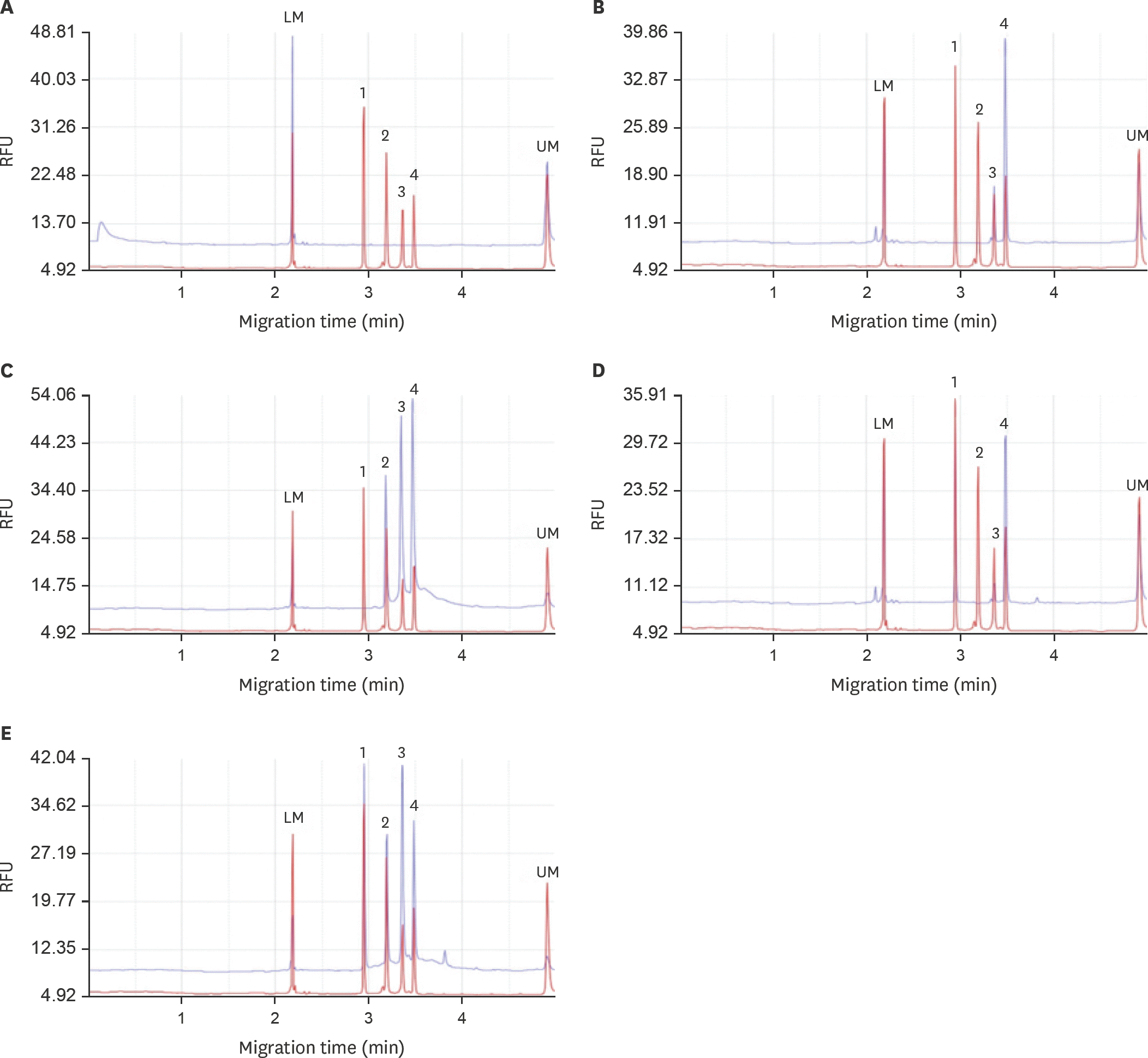 | Fig. 2.The representative capillary electrophoresis results of the field samples (Blue line, tested field sample; Red line, positive control which was from the in vitro transcribed RNA standards). (A) All negative. (B) NP and H6 positive. (C) NP, H6 and H5 positive. (D) NP, H6 and H7 positive. (E) All of the four genes (NP, H6, H5, and H7) were positive. LM, lower alignment marker (20 bp); UM, upper alignment marker (1,000 bp); Peak 1, H7 HA gene (137 bp); Peak 2, H5 HA gene (173 bp); Peak 3, H6 HA gene (199 bp); Peak 4, NP gene (217 bp); NP, nucleoprotein. |
Table 1.
The designed recombinase polymerase amplification primers for multiple detection of AIV genes




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download