Abstract
Several gut commensals have been shown to modulate host immune response. Recently, many food derived microbes have also been reported to affect the immune system. However, a mechanism to identify immunostimulatory and immunoregulatory microbes is needed. Here, we successfully established an in vitro screening system and identified an immunoregulatory bacterium, Lactobacillus pentosus KF340 (LP340), present in various fermented foods. LP340 induced a regulatory phenotype in mice Ag presenting cells which, in turn, induced IL-10 and IFN-γ producing Type 1 regulatory T cells (Tr1 cells) from naïve CD4+ T cells. Naïve CD4+ T cells co-cultured with LP340 treated dendritic cells highly expressed cytokine receptor IL-27R and were CD49b and lymphocyte-activation gene 3 double positive. Oral administration of LP340 in mice with atopic dermatitis reduced cellular infiltration in affected ear lobes and serum IgE levels, thus, ameliorating the disease symptoms. This suggests a systemic immunoregulatory effect of LP340. These findings demonstrate that LP340, a bacterium derived from food, prevents systemic inflammation through the induction of IL-10 producing Tr1 cells.
Go to : 
References
1. Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. 2016; 352:539–544.

2. Mosconi I, Geuking MB, Zaiss MM, Massacand JC, Aschwanden C, Kwong Chung CK, McCoy KD, Harris NL. Intestinal bacteria induce TSLP to promote mutualistic T-cell responses. Mucosal Immunol. 2013; 6:1157–1167.

3. Gomez de Agüero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, Steinert A, Heikenwalder M, Hapfelmeier S, Sauer U, et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016; 351:1296–1302.

4. Kwon HK, Lee CG, So JS, Chae CS, Hwang JS, Sahoo A, Nam JH, Rhee JH, Hwang KC, Im SH. Generation of regulatory dendritic cells and CD4+Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc Natl Acad Sci U S A. 2010; 107:2159–2164.

5. Abdel-Gadir A, Stephen-Victor E, Gerber GK, Noval Rivas M, Wang S, Harb H, Wang L, Li N, Crestani E, Spielman S, et al. Microbiota therapy acts via a regulatory T cell MyD88/RORγ t pathway to suppress food allergy. Nat Med. 2019; 25:1164–1174.

6. Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018; 555:210–215.

7. Coqueiro AY, Raizel R, Bonvini A, Tirapegui J, Rogero MM. Probiotics for inflammatory bowel diseases: a promising adjuvant treatment. Int J Food Sci Nutr. 2019; 70:20–29.

8. He B, Hoang TK, Tian X, Taylor CM, Blanchard E, Luo M, Bhattacharjee MB, Freeborn J, Park S, Couturier J, et al. Lactobacillus reuteri reduces the severity of experimental autoimmune encephalomyelitis in mice by modulating gut microbiota. Front Immunol. 2019; 10:385.

9. Liu X, Zeng B, Zhang J, Li W, Mou F, Wang H, Zou Q, Zhong B, Wu L, Wei H, et al. Role of the gut microbiome in modulating arthritis progression in mice. Sci Rep. 2016; 6:30594.

10. Ng QX, Loke W, Venkatanarayanan N, Lim DY, Soh AYS, Yeo WS. A systematic review of the role of prebiotics and probiotics in autism spectrum disorders. Medicina (Kaunas). 2019; 55:E129.

11. Zeng H, Zhang R, Jin B, Chen L. Type 1 regulatory T cells: a new mechanism of peripheral immune tolerance. Cell Mol Immunol. 2015; 12:566–571.

12. Gagliani N, Magnani CF, Huber S, Gianolini ME, Pala M, Licona-Limon P, Guo B, Herbert DR, Bulfone A, Trentini F, et al. Coexpression of CD49b and LAG-3 identifies human and mouse T regulatory type 1 cells. Nat Med. 2013; 19:739–746.

13. Chihara N, Madi A, Karwacz K, Awasthi A, Kuchroo VK. Differentiation and characterization of Tr1 cells. Curr Protoc Immunol. 2016; 113:3. 27.1–3.27.10.

14. Biedermann T, Skabytska Y, Kaesler S, Volz T. Regulation of T cell immunity in atopic dermatitis by microbes: the yin and yang of cutaneous inflammation. Front Immunol. 2015; 6:353.

15. Mayo L, Cunha AP, Madi A, Beynon V, Yang Z, Alvarez JI, Prat A, Sobel RA, Kobzik L, Lassmann H, et al. IL-10-dependent Tr1 cells attenuate astrocyte activation and ameliorate chronic central nervous system inflammation. Brain. 2016; 139:1939–1957.

16. Bae MJ, Kim HK, Lim S, Lee SY, Shin HS, Kim JE, Im SH, Kim S. Lactobacillus pentosus KF340 alleviates house dust mite-induced murine atopic dermatitis via the secretion of il-10-producing splenic b10 cells. J Funct Foods. 2016; 26:258–267.
17. Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu Rev Immunol. 2011; 29:71–109.

18. Tait Wojno ED, Hunter CA, Stumhofer JS. The immunobiology of the interleukin-12 family: room for discovery. Immunity. 2019; 50:851–870.

19. Teng MW, Bowman EP, McElwee JJ, Smyth MJ, Casanova JL, Cooper AM, Cua DJ. IL-12 and IL-23 cytokines: from discovery to targeted therapies for immune-mediated inflammatory diseases. Nat Med. 2015; 21:719–729.

20. Vignali DA, Kuchroo VK. IL-12 family cytokines: immunological playmakers. Nat Immunol. 2012; 13:722–728.

21. Funderburg NT, Jadlowsky JK, Lederman MM, Feng Z, Weinberg A, Sieg SF. The Toll-like receptor 1/2 agonists Pam(3) CSK(4) and human β-defensin-3 differentially induce interleukin-10 and nuclear factor-κ B signalling patterns in human monocytes. Immunology. 2011; 134:151–160.
22. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015; 350:1084–1089.
23. Li H, Shi B. Tolerogenic dendritic cells and their applications in transplantation. Cell Mol Immunol. 2015; 12:24–30.

24. Laidlaw BJ, Cui W, Amezquita RA, Gray SM, Guan T, Lu Y, Kobayashi Y, Flavell RA, Kleinstein SH, Craft J, et al. Production of IL-10 by CD4 (+) regulatory T cells during the resolution of infection promotes the maturation of memory CD8 (+) T cells. Nat Immunol. 2015; 16:871–879.

25. Chaudhry A, Samstein RM, Treuting P, Liang Y, Pils MC, Heinrich JM, Jack RS, Wunderlich FT, Brüning JC, Müller W, et al. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity. 2011; 34:566–578.

26. Brockmann L, Soukou S, Steglich B, Czarnewski P, Zhao L, Wende S, Bedke T, Ergen C, Manthey C, Agalioti T, et al. Molecular and functional heterogeneity of IL-10-producing CD4+ T cells. Nat Commun. 2018; 9:5457.

27. Andolfi G, Fousteri G, Rossetti M, Magnani CF, Jofra T, Locafaro G, Bondanza A, Gregori S, Roncarolo MG. Enforced IL-10 expression confers type 1 regulatory T cell (Tr1) phenotype and function to human CD4 (+) T cells. Mol Ther. 2012; 20:1778–1790.

28. Iwasaki Y, Fujio K, Okamura T, Yanai A, Sumitomo S, Shoda H, Tamura T, Yoshida H, Charnay P, Yamamoto K. Egr-2 transcription factor is required for Blimp-1-mediated IL-10 production in IL-27-stimulated CD4+ T cells. Eur J Immunol. 2013; 43:1063–1073.
29. Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008; 453:620–625.

30. Verma R, Lee C, Jeun EJ, Yi J, Kim KS, Ghosh A, Byun S, Lee CG, Kang HJ, Kim GC, et al. Cell surface polysaccharides of Bifidobacterium bifidum induce the generation of foxp3+ regulatory T cells. Sci Immunol. 2018; 3:eaat6975.

31. Collins FL, Rios-Arce ND, Schepper JD, Jones AD, Schaefer L, Britton RA, McCabe LR, Parameswaran N. Beneficial effects of Lactobacillus reuteri 6475 on bone density in male mice is dependent on lymphocytes. Sci Rep. 2019; 9:14708.

32. Jia H, Ren S, Wang X. Heat-killed probiotic regulates the body's regulatory immunity to attenuate subsequent experimental autoimmune arthritis. Immunol Lett. 2019; 216:89–96.

33. Lim SK, Kwon MS, Lee J, Oh YJ, Jang JY, Lee JH, Park HW, Nam YD, Seo MJ, Roh SW, et al. Weissella cibaria WIKIM28 ameliorates atopic dermatitis-like skin lesions by inducing tolerogenic dendritic cells and regulatory T cells in BALB/c mice. Sci Rep. 2017; 7:40040.

34. Jeon SG, Kayama H, Ueda Y, Takahashi T, Asahara T, Tsuji H, Tsuji NM, Kiyono H, Ma JS, Kusu T, et al. Probiotic Bifidobacterium breve induces IL-10-producing Tr1 cells in the colon. PLoS Pathog. 2012; 8:e1002714.
35. Jonuleit H, Schmitt E, Schuler G, Knop J, Enk AH. Induction of interleukin 10-producing, nonproliferating CD4 (+) T cells with regulatory properties by repetitive stimulation with allogeneic immature human dendritic cells. J Exp Med. 2000; 192:1213–1222.

36. Gregori S, Tomasoni D, Pacciani V, Scirpoli M, Battaglia M, Magnani CF, Hauben E, Roncarolo MG. Differentiation of type 1 T regulatory cells (Tr1) by tolerogenic DC-10 requires the IL-10-dependent ILT4/HLA-G pathway. Blood. 2010; 116:935–944.

37. Wang H, Meng R, Li Z, Yang B, Liu Y, Huang F, Zhang J, Chen H, Wu C. IL-27 induces the differentiation of Tr1-like cells from human naive CD4+ T cells via the phosphorylation of STAT1 and STAT3. Immunol Lett. 2011; 136:21–28.

38. Vasanthakumar A, Kallies A. IL-27 paves different roads to Tr1. Eur J Immunol. 2013; 43:882–885.

39. Boonstra A, Rajsbaum R, Holman M, Marques R, Asselin-Paturel C, Pereira JP, Bates EE, Akira S, Vieira P, Liu YJ, et al. Macrophages and myeloid dendritic cells, but not plasmacytoid dendritic cells, produce IL-10 in response to MyD88- and TRIF-dependent TLR signals, and TLR-independent signals. J Immunol. 2006; 177:7551–7558.

40. Villagra A, Cheng F, Wang HW, Suarez I, Glozak M, Maurin M, Nguyen D, Wright KL, Atadja PW, Bhalla K, et al. The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance. Nat Immunol. 2009; 10:92–100.

41. Hu X, Paik PK, Chen J, Yarilina A, Kockeritz L, Lu TT, Woodgett JR, Ivashkiv LB. IFN-gamma suppresses IL-10 production and synergizes with TLR2 by regulating GSK3 and CREB/AP-1 proteins. Immunity. 2006; 24:563–574.
42. Geijtenbeek TB, Van Vliet SJ, Koppel EA, Sanchez-Hernandez M, Vandenbroucke-Grauls CM, Appelmelk B, Van Kooyk Y. Mycobacteria target DC-SIGN to suppress dendritic cell function. J Exp Med. 2003; 197:7–17.

43. Rogers NC, Slack EC, Edwards AD, Nolte MA, Schulz O, Schweighoffer E, Williams DL, Gordon S, Tybulewicz VL, Brown GD, et al. Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins. Immunity. 2005; 22:507–517.

44. Dennehy KM, Willment JA, Williams DL, Brown GD. Reciprocal regulation of IL-23 and IL-12 following co-activation of Dectin-1 and TLR signaling pathways. Eur J Immunol. 2009; 39:1379–1386.

45. Edwards AD, Manickasingham SP, Spörri R, Diebold SS, Schulz O, Sher A, Kaisho T, Akira S, Reis e Sousa C. Microbial recognition via Toll-like receptor-dependent and -independent pathways determines the cytokine response of murine dendritic cell subsets to CD40 triggering. J Immunol. 2002; 169:3652–3660.

46. Caruso R, Warner N, Inohara N, Núñez G. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity. 2014; 41:898–908.

47. Neuper T, Ellwanger K, Schwarz H, Kufer TA, Duschl A, Horejs-Hoeck J. NOD1 modulates IL-10 signalling in human dendritic cells. Sci Rep. 2017; 7:1005.

48. Patin EC, Jones AV, Thompson A, Clement M, Liao CT, Griffiths JS, Wallace LE, Bryant CE, Lang R, Rosenstiel P, et al. IL-27 induced by select candida spp. Via TLR7/NOD2 signaling and IFN-beta production inhibits fungal clearance. J Immunol. 2016; 197:208–221.
49. Hawrylowicz CM. Regulatory T cells and IL-10 in allergic inflammation. J Exp Med. 2005; 202:1459–1463.

50. Huang W, Solouki S, Koylass N, Zheng SG, August A. ITK signalling via the Ras/IRF4 pathway regulates the development and function of Tr1 cells. Nat Commun. 2017; 8:15871.

51. Boyman O, Werfel T, Akdis CA. The suppressive role of IL-10 in contact and atopic dermatitis. J Allergy Clin Immunol. 2012; 129:160–161.

52. Volz T, Skabytska Y, Guenova E, Chen KM, Frick JS, Kirschning CJ, Kaesler S, Röcken M, Biedermann T. Nonpathogenic bacteria alleviating atopic dermatitis inflammation induce IL-10-producing dendritic cells and regulatory Tr1 cells. J Invest Dermatol. 2014; 134:96–104.

53. Zhang H, Kong H, Zeng X, Guo L, Sun X, He S. Subsets of regulatory T cells and their roles in allergy. J Transl Med. 2014; 12:125.

54. Kennedy Norton S, Barnstein B, Brenzovich J, Bailey DP, Kashyap M, Speiran K, Ford J, Conrad D, Watowich S, Moralle MR, et al. IL-10 suppresses mast cell IgE receptor expression and signaling in vitro and in vivo. J Immunol. 2008; 180:2848–2854.
Go to : 
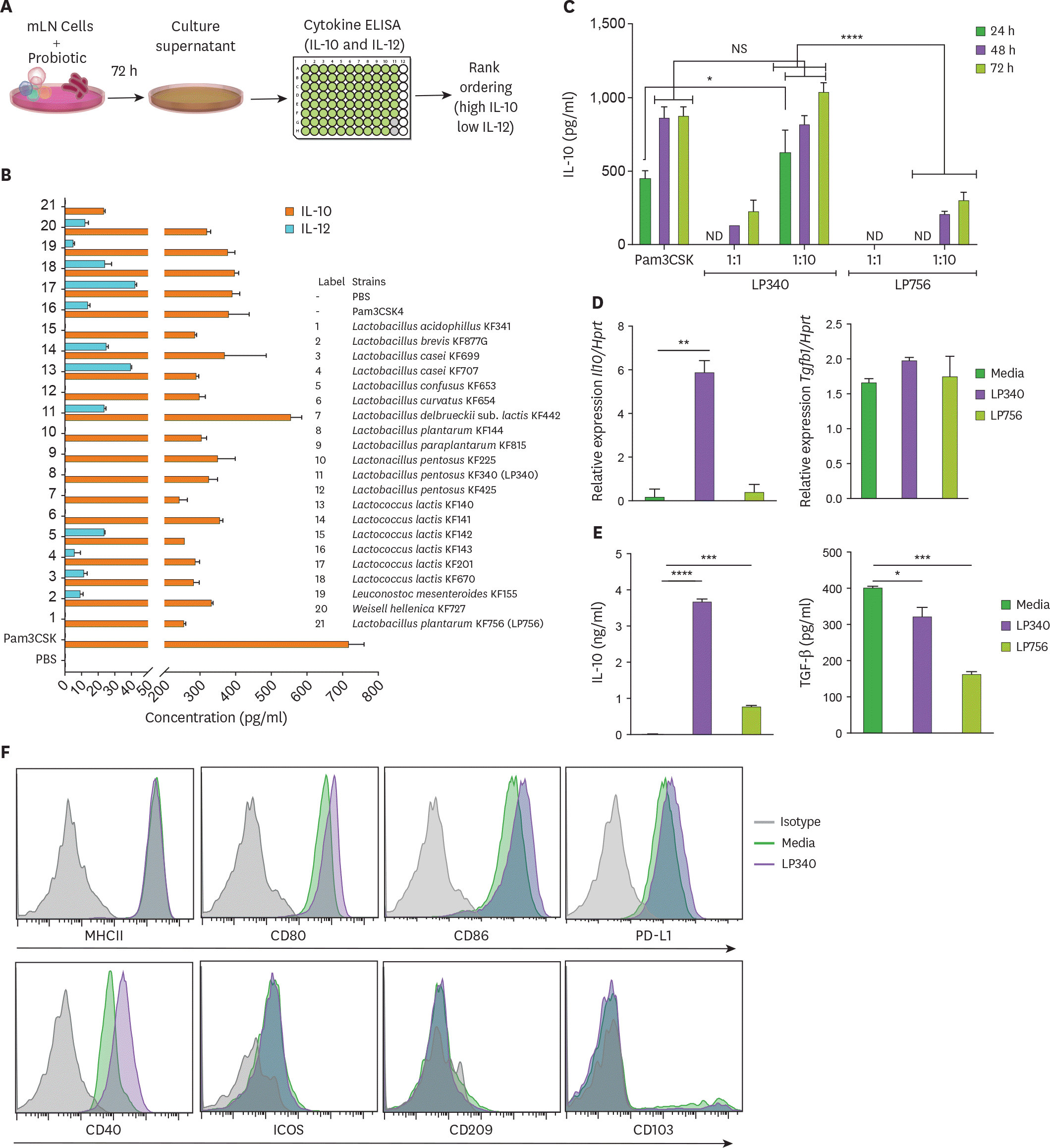 | Figure 1.LP340 preferentially skews the immune microenvironment to immunoregulatory phenotype, both with total mLN cells and APCs. (A-C) Total mLN cells were co-cultured with probiotics and supernatant was used for ELISA, as described in (A) schematics of primary screening method (B) cytokines, IL-10 and IL-12 levels (C) cells were treated with LP340 and LP756 at different concentrations and incubation times followed by measurement of cytokines. (D-F) enriched splenic CD11c+ cells were treated with LP340 or LP756 and (D) qRT-PCR analysis of Il10 and Tgfb1 gene expression (E) cytokine analysis by ELISA and (F) flow cytometry analysis of MHCII, CD80, CD86, PD-L1, CD40, ICOS, CD209, and CD103. All data are representative of 3 independent experiments and are presented as mean±SD. NS, not significant; ND, not detectable; ICOS, inducible T-cell costimulator. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001. |
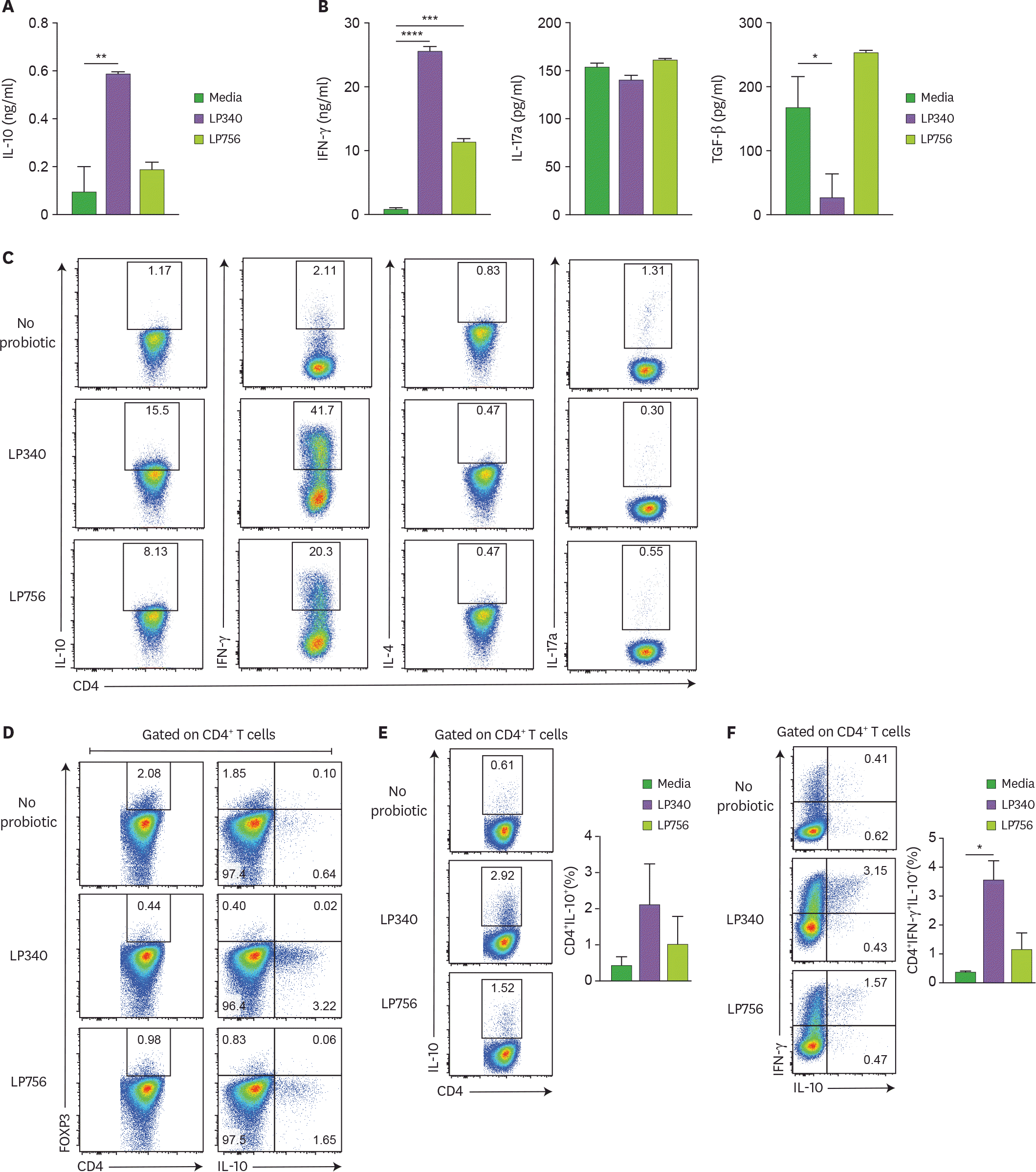 | Figure 2.DCs exposed to LP340 induce CD4+ T-cells to produce IL-10. (A-D) Splenic CD11c+ cells were co-cultured with LP340 and LP756, then primed CD11c+ cells were co-cultured with naïve CD4+ T-cells in presence of anti-CD3 and IL-2. Culture supernatants were used for ELISA of cytokines (A) IL-10 (B) IFN-γ, IL-17A, and TGF-β. (C) CD4+ T-cells were stimulated with PMA/Ionomycin and used for flow cytometry analysis of cytokines IL-10, IFN-γ, IL-4 and IL-17A (D) CD4+ T-cells were also analyzed by flow cytometry for FOXP3 expression and IL-10 expression in CD4+ T-cells. (E) CD4+ IL-10+, and (F) CD4+ IFN-γ+ IL-10+ T-cells were analyzed by flow cytometry. All data are representative of 3 independent experiments and presented as mean±SD. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001. |
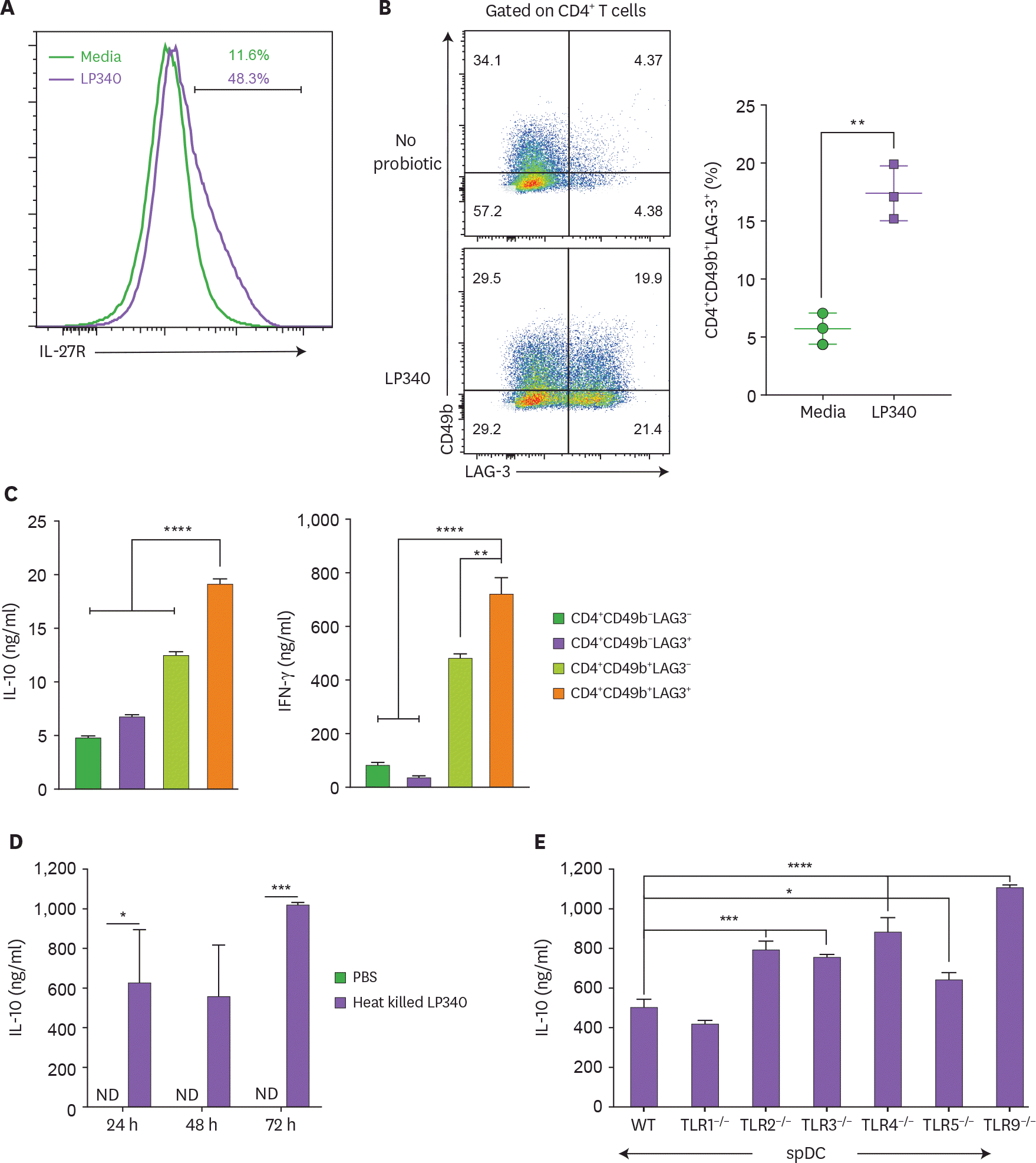 | Figure 3.LP340 induce a Tr-1 cell like phenotype in CD4+ T-cells. (A-E) Splenic CD11c+ cells were co-cultured with LP340 and primed CD11c+ cells were co-cultured with naïve CD4+ T-cells in presence of anti-CD3 and IL-2. (A) Cells were analyzed for IL-27R and (B) CD49b+ LAG-3+ expression, gated on total CD4+ T-cells. The 4 populations sorted by expression of CD49b & LAG-3 were cultured and cell supernatants were used for (C) IL-10 & IFN-γ cytokine ELISA (D) Heat killed LP340 was used for priming the DCs and IL-10 ELISA was performed with supernatant (E) Splenic CD11c+ cells from TLR knock-out mice (TLR1−/−, TLR2−/−, TLR3−/−, TLR4−/−, TLR5−/−, and TLR9−/−) were treated with LP340 and IL-10 cytokine ELISA was performed. Data are representative of 3 (A-D) and 2 (E) independent experiments and presented as mean±SD. IL-27R, IL-27 receptor; ND, not detectable; WT, wild type. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001. |
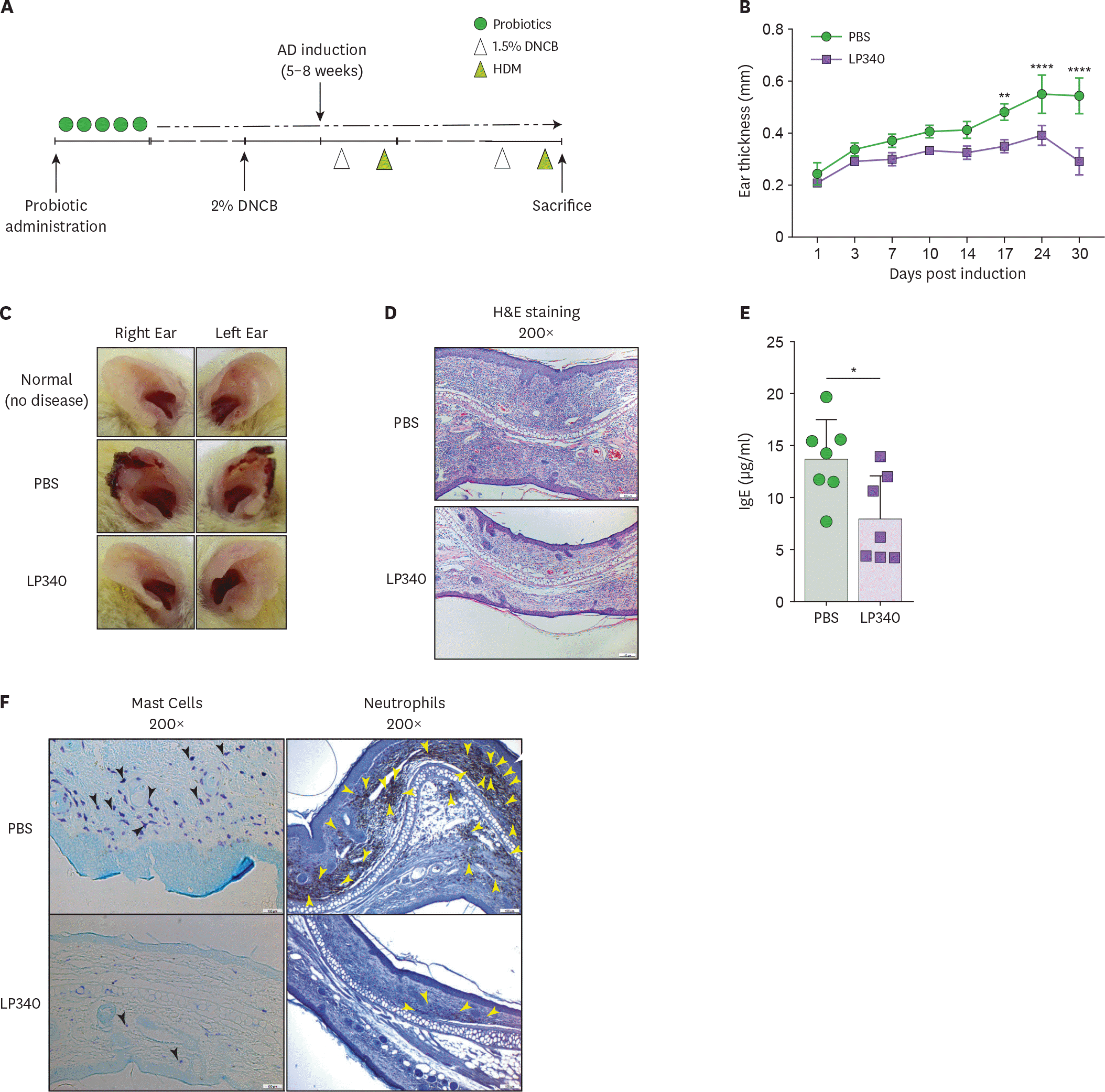 | Figure 4.LP340 ameliorates HDM induced AD. (A-F) Mice orally fed with PBS or LP340 were immunized with DNCB and subsequently were challenged with HDM as depicted in (A) experimental scheme. (B) Ear thickness was measured till 30 days post induction and (C) gross ear lesions were observed (D) Inflammation and cellular infiltration in ear tissue was analyzed by histology, H&E staining (E) Serum IgE levels were measured by ELISA (F) Mast cell (left panel, black arrowheads) and neutrophil (right panel, yellow arrowheads) infiltration was observed by IHC of ear tissue. Data are representative of 3 independent experiments and presented as mean±SD. * p<0.05, ** p<0.01, **** p<0.0001. |
Table 1.
Primer sequences for qRT-PCR




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download