1. Koo BK, Park S, Han KD, Moon MK. Hypertriglyceridemia is an independent risk factor for cardiovascular diseases in Korean adults aged 30–49 years: a nationwide population-based study. J Lipid Atheroscler. 2021; 10:88–98. PMID:
33537256.

2. Cho SM, Lee H, Lee HH, et al. Dyslipidemia fact sheets in Korea 2020: an analysis of nationwide population-based data. J Lipid Atheroscler. 2021; 10:202–209. PMID:
34095012.

3. McPherson R, Tybjaerg-Hansen A. Genetics of coronary artery disease. Circ Res. 2016; 118:564–578. PMID:
26892958.

4. Lee CJ, Lee JH, Choi S, et al. Screening, diagnosis, and treatment of familial hypercholesterolemia: symposium of the education committee, Korean Society of Lipid and Atherosclerosis. J Lipid Atheroscler. 2018; 7:122–154.

5. Mach F, Baigent C, Catapano AL, et al. 2019 ESC/EAS guidelines for the management of dyslipidaemias: lipid modification to reduce cardiovascular risk. Eur Heart J. 2020; 41:111–188. PMID:
31504418.
6. Han SM, Hwang B, Park TG, et al. Genetic testing of Korean familial hypercholesterolemia using whole-exome sequencing. PLoS One. 2015; 10:e0126706. PMID:
25962062.

7. Yamashita S, Sprecher DL, Sakai N, Matsuzawa Y, Tarui S, Hui DY. Accumulation of apolipoprotein E-rich high density lipoproteins in hyperalphalipoproteinemic human subjects with plasma cholesteryl ester transfer protein deficiency. J Clin Invest. 1990; 86:688–695. PMID:
2118552.

8. Lee CJ, Lee Y, Park S, et al. Rare and common variants of APOB and PCSK9 in Korean patients with extremely low low-density lipoprotein-cholesterol levels. PLoS One. 2017; 12:e0186446. PMID:
29036232.

9. Lee CJ, Oum CY, Lee Y, et al. Variants of lipolysis-related genes in Korean patients with very high triglycerides. Yonsei Med J. 2018; 59:148–153. PMID:
29214790.

10. Lee CJ, Park MS, Kim M, et al. CETP, LIPC, and SCARB1 variants in individuals with extremely high high-density lipoprotein-cholesterol levels. Sci Rep. 2019; 9:10915. PMID:
31358896.

11. Tada H, Kawashiri MA, Nohara A, Inazu A, Mabuchi H, Yamagishi M. Impact of clinical signs and genetic diagnosis of familial hypercholesterolaemia on the prevalence of coronary artery disease in patients with severe hypercholesterolaemia. Eur Heart J. 2017; 38:1573–1579. PMID:
28159968.
12. Santos PC, Morgan AC, Jannes CE, et al. Presence and type of low density lipoprotein receptor (LDLR) mutation influences the lipid profile and response to lipid-lowering therapy in Brazilian patients with heterozygous familial hypercholesterolemia. Atherosclerosis. 2014; 233:206–210. PMID:
24529145.

13. Santos PC, Pereira AC. Type of LDLR mutation and the pharmacogenetics of familial hypercholesterolemia treatment. Pharmacogenomics. 2015; 16:1743–1750. PMID:
26427613.

14. Perez de Isla L, Alonso R, Watts GF, et al. Attainment of LDL-cholesterol treatment goals in patients with familial hypercholesterolemia: 5-year SAFEHEART registry follow-up. J Am Coll Cardiol. 2016; 67:1278–1285. PMID:
26988947.
15. Kim H, Lee CJ, Pak H, et al. GENetic characteristics and REsponse to lipid-lowering therapy in familial hypercholesterolemia: GENRE-FH study. Sci Rep. 2020; 10:19336. PMID:
33168860.

16. Postmus I, Trompet S, Deshmukh HA, et al. Pharmacogenetic meta-analysis of genome-wide association studies of LDL cholesterol response to statins. Nat Commun. 2014; 5:5068. PMID:
25350695.
17. Natarajan P, Young R, Stitziel NO, et al. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation. 2017; 135:2091–2101. PMID:
28223407.

18. Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010; 466:707–713. PMID:
20686565.
19. Crosby J, Peloso GM, Auer PL, et al. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N Engl J Med. 2014; 371:22–31. PMID:
24941081.

20. Spiller W, Jung KJ, Lee JY, Jee SH. Precision medicine and cardiovascular health: insights from Mendelian randomization analyses. Korean Circ J. 2020; 50:91–111. PMID:
31845553.

21. Lee SH, Lee JY, Kim GH, et al. Two-sample Mendelian randomization study of lipid levels and ischemic heart disease. Korean Circ J. 2020; 50:940–948. PMID:
32812402.

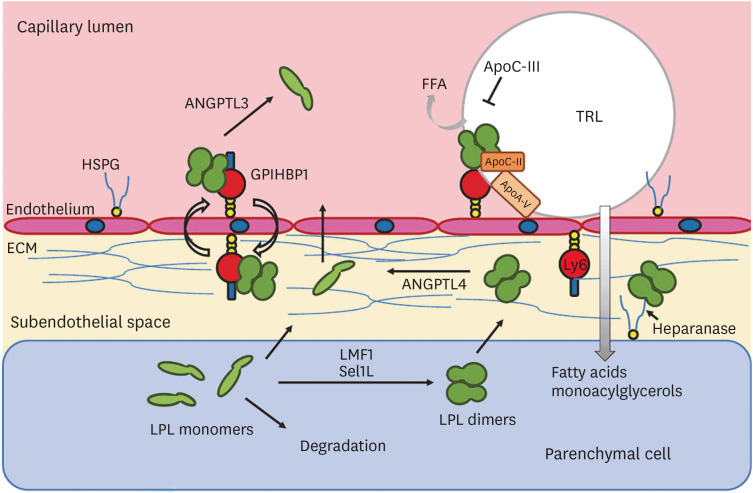
22. Olivecrona G. Role of lipoprotein lipase in lipid metabolism. Curr Opin Lipidol. 2016; 27:233–241. PMID:
27031275.

23. Musunuru K, Pirruccello JP, Do R, et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med. 2010; 363:2220–2227. PMID:
20942659.
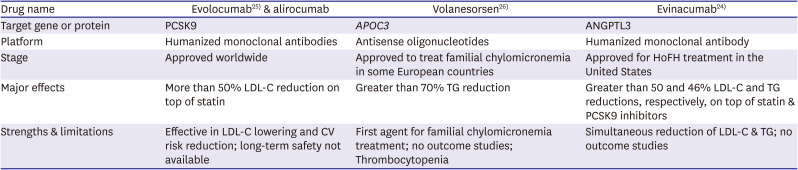
24. Dewey FE, Gusarova V, O'Dushlaine C, et al. Inactivating variants in ANGPTL4 and risk of coronary artery disease. N Engl J Med. 2016; 374:1123–1133. PMID:
26933753.

25. Levin AA. Treating disease at the RNA level with oligonucleotides. N Engl J Med. 2019; 380:57–70. PMID:
30601736.

26. Raal FJ, Rosenson RS, Reeskamp LF, et al. Evinacumab for homozygous familial hyperchoelsterolemia. N Engl J Med. 2020; 383:711–720. PMID:
32813947.
27. Sabatine MS, Giugliano RP, Keech AC, et al. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med. 2017; 376:1713–1722. PMID:
28304224.

28. Witztum JL, Gaudet D, Freedman SD, et al. Volanesorsen and triglyceride levels in familial chylomicronemia syndrome. N Engl J Med. 2019; 381:531–542. PMID:
31390500.





 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download