Introduction
Methods
Table 1
Study selection
Data extraction and synthesis
Risk of bias assessment
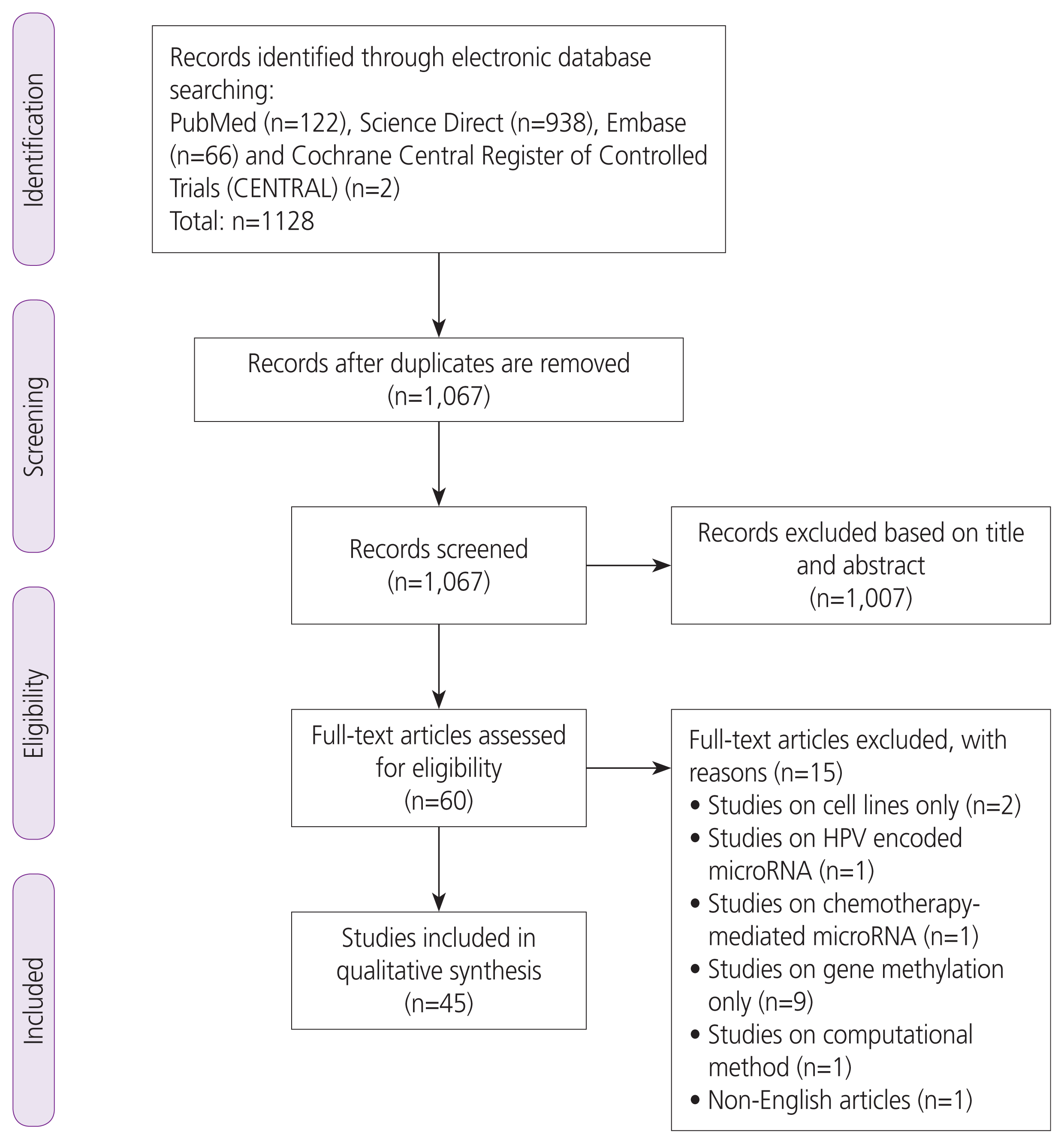
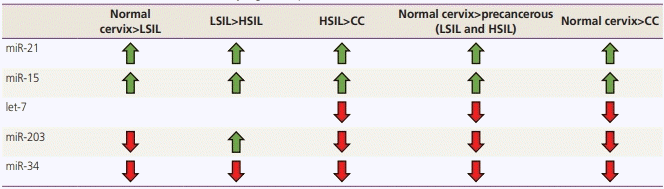
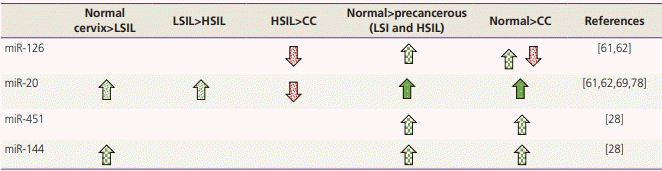
Results
Characteristics of selected studies
Table 2
| Title | Author, year, country, type of study | Sample size, sample type | Age group | Method, reference gene | miRNA mentioned (significant) | Normal cervix to CC | Normal cervix to CIN I-III | Normal cervix to LSIL (CIN I) | LSIL (CIN I) to HSIL (CIN II-III) | HSIL (CIN II-III) to CC | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|||||||||||
| Upregulated | Downregulated | Upregulated | Downregulated | Upregulated | Downregulated | Upregulated | Downregulated | Upregulated | Downregulated | ||||||
| 1. Oncogenic microRNA signature for early diagnosis of cervical intraepithelial neoplasia and cancer | Liu et al. [39], 2018, Hong Kong, case-control study | 145 NILM, 239 LSIL, 285 HSIL, 58 CC, frozen and FFPE tissue | Mean: NILM (50), LSIL (39), HSIL (42), CC (51) | Microarray analysis and validated by RT-qPCR, RNU6B | Mir-20a, Mir-92a, Mir-141, Mir-183, Mir-210, Mir-944 | Mir-20a, Mir-92a, Mir-141, Mir-183, Mir-210, Mir-944 | Mir-20a, Mir-92a, Mir-141, Mir-183, Mir-210, Mir-944 | Mir-20a, Mir-92a, Mir-141, Mir-183, Mir-210, Mir-944 | Mir-92a, Mir-183 | Mir-20a, Mir-141, Mir-210, Mir-944 | |||||
|
|
|||||||||||||||
| 2. Microrna-551b expression profile in low and high-grade cervical intraepithelial neoplasia | Lukic et al. [30], 2018, Italy, case-control study | 10 Normal, 18 condyloma, 8 CIN I, 14 CIN II-III, FFPE tissue | Mean: 35.98±9 | RT-qPCR, U6snRNA | Mir-551b | Mir-551b | Mir-551b | Mir-551b | |||||||
|
|
|||||||||||||||
| 3. Mir-155-5p inhibits PDK1 and promotes autophagy via the mtor pathway in cervical cancer | Wang et al. [43], 2018, China, case-control study | 48 Normal, 18 CIN I-II, 18 CIN III-CC, fresh tissue | N/A | RT-qPCR, U6snRNA | Mir-155-5p | ||||||||||
|
|
|||||||||||||||
| 4. Comparable expression of Mir-Let-7b, Mir-21, Mir-182, Mir-145, and P53 in serum and cervical cells: diagnostic implications for early detection of cervical lesions | Okoye et al. [35], 2019, Nigeria, cross-sectional study | 159 NILM, 46 cervicitis, 46 ASCUS, 40 LSIL, 28 HSIL, 10 SCC, serum and exfoliated cells | Mean: 39.45±11.16 | PCR and gel electrophoresis, Mir-16 | Mir-21, Mir-146a, Mir-155, Mir-200c, Mir-182, Let-7b, Mir-145 | Mir-21, Mir-146a, Mir-155, Mir-200c | Mir-21, Mir-146a, Mir-182 | Mir-145 | Mir-182, Mir-21, Mir-182, Mir-200c (cell) | Mir-146a, Mir-200c (serum) | Mir-21, Mir-146a, Mir-155, Mir-200, Let-7b | Mir-182 | |||
|
|
|||||||||||||||
| 5. MicroRNA expressions in HPV-Induced cervical dysplasia and cancer | Gocze et al. [31], 2015, Hungary, cross-sectional study | 30 CINI, 10 CIN, 20 CIN III, 38 SCC, FFPE tissue | Mean: 36.17 | RT-qPCR, 5srRNA, U6snRNA | Mir-27a, Mir-34a, Mir-155, Mir-196a, Mir-203 | Mir-27a, Mir-155 | Mir-34a | Mir-27a | Mir-196a, Mir-34a | ||||||
|
|
|||||||||||||||
| 6. Plasma expression of mirna-21, -214, -34a, and -200a in patients with persistent HPV infection and cervical lesions | Yang et al. [45], 2019, China, case-control study | 73 Normal, 19 CIN I, 54 CIN II, 71 CIN III, 15 CC, serum and FFPE tissue | Mean: normal (40.94±7.23), CIN1 (41.93±9.10), CIN2 (42.37±9.71), CIN3 (40.77±8.09), CC (49.73±8.42) | RT-qPCR, Mir-Let-7 | miRNA-21, Mir-214, Mir-34a, Mir-200a | Mir-21 | Mir-214, Mir-200a, Mir -34a | Mir-214, Mir-200a, Mir -34a | |||||||
|
|
|||||||||||||||
| 7. Microrna-466 with tumour markers for cervical cancer screening | Zhou et al. [44], 2017, China, cohort study | 210 Normal, 280 cervical hyperplasia, 143 SCC, 102 AD, serum | Median: 49 | RT-qPCR, U6snRNA | Mir-466 | Mir-466 | |||||||||
|
|
|||||||||||||||
| 8. Expression profile of_microRNA-203_and Its__np63 target In cervical carcinogenesis: prospects for cervical cancer screening | Piersma et al. [34], 2016, Brazil, cross-sectional study | 21 Normal, 18 CIN I, 20 CIN II, 21 CIN III, 15 CC, fresh tissue | N/A | RT-qPCR, Mie-191, Mir-23a | Mir-203 | Mir-203 | Mir-203 | Mir-203 | Mir-203 | ||||||
|
|
|||||||||||||||
| 9. Novel microRNA signatures in HPV-mediated cervical carcinogenesis in Indian women | Sharma et al. [23], 2016, India, case-control study | 30 Normal, 20 CIN, 50 CC, frozen tissue | N/A | RT-qPCR, U6 snRNA, RNU43, RtU snRNA | Mir-200b, Mir-146a, Mir-3130-5p, Mir-95, Mir-136, Mir-34a, Let-7a, Mir-23b, Mir-29, Mir-19a, Mir-222, Mir-1, Mir-29a, Mir-576-3, Mir-21, Mir-449b, Mir-184, Mir-17, Mir-99a, Mir-376b, Mir-193-5p, Mir-423-5p, Mir-15a, Mir-517a, Mir-545, Mir-223, Mir-192 | Mir-15a, Mir-449b, Mir-517a, Mir-545, Mir-223, Mir-192 | Mir-34a, Mir-23b, Mir-99a, Mir-376b, Mir-193-5p, Mir-423-5p | Mir-29, Mir-19a, Mir-222, Mir-1, Mir-29a, Mir-222 and Mir-19a, Mir-576-3, Mir-21, Mir-449b, Mir-184, Mir-17 | Mir-200b, Mir-146a, Mir-3130-5p, Mir-95 and Mir-136, Mir-34a, Let-7a, Mir-23b | ||||||
|
|
|||||||||||||||
| 10. Up-regulation of mir-21 is associated with cervicitis and human papillomavirus infection in cervical tissues | Bumrungthai et al. [27], 2015, Thailand, cross-sectional study | 20 Normal, 12 cervicitis, 14 CIN I, 22 CIN II-III, and 43 SCC, fresh tissue and exfoliated cells | N/A | RT-qPCR, U6snRNA | Mir-21 | Mir-21 | Mir-21 | ||||||||
|
|
|||||||||||||||
| 11. Mir-34a and mir-125b expression in HPV infection and cervical cancer development | Ribeiro et al. [20], 2015, Portugal, cross-sectional study | 49 Normal, 28 LSIL, 29 HSIL, 8 ICC, exfoliated cells | Mean: 40±12.6 | RT-qPCR, Mir-23a | Mir-34a, Mir-125b | Mir-34a | Mir-125b | ||||||||
|
|
|||||||||||||||
| 12. Detection of high-grade neoplasia in air-dried cervical PAP smears by a microRNA-based classifier | Ivanov et al. [29], 2018, Russia, cross-sectional study | 40 Normal, 34 LSIL, 57 HSIL, 43 ICC, exfoliated cells | Mean: normal (31), LSIL (36), HSIL (44), ICC (53) | RT-qPCR, U6snRNA | Mir-106b, Mir-1246, Mir-126, Mir-196b, Mir-20a, Mir-21, Mir-375, Mir-145 | Mir-375, Mir-145 | Mir-106, Mir-1246, Mir-126, Mir-196b, Mir-20a, Mir-21 | Mir-375, Mir-145 | Mir-106b, Mir-1246, Mir-126, Mir-196b, Mir-20a, Mir-21 | ||||||
|
|
|||||||||||||||
| 13. Mir-21-5p, Mir-34a, and human telomerase RNA component as surrogate markers for cervical cancer progression | Zhu et al. [53], 2018, China, case-control study | 18 NILM, 45 ASCUS, 21 LSIL, 25 HSIL, 9 SCC, fresh tissue and exfoliated cells | Mean: 43.5±8.5 | RT-qPCR, U6snRNA | Mir-21-5p, Mir-34a | Mir-21-5p, | Mir-34a | Mir-21-5p | Mir-34a | Mir-21-5p | Mir-34a | Mir-21-5p | Mir-34a | Mir-21-5p | Mir-34a |
|
|
|||||||||||||||
| 14. Identification of miRNAs in cervical mucus as A novel diagnostic marker for cervical neoplasia | Kawai et al. [28], 2018, Japan, cross-sectional study | 56 Normal, 19 CIN I, 33 CIN II, 43 CIN III, 35 SCC, 19 AD, mucus and exfoliated cells | Median: normal (36), CIN I (39), CIN II (38), CIN III (36), SCC (54), AD (47) | Microarray analysis and validated by RT-qPCR, RNU48 | Mir-126-3p, Mir -20b-5p, Mir-451a, Mir-144-3p | Mir-126-3p, Mir-20b-5p, Mir-451a, Mir-144-3p | Mir-126-3p, Mir-20b-5p, Mir-451a, Mir-144-3p | Mir-144-3p | |||||||
|
|
|||||||||||||||
| 15. Triage of high-risk HPV-positive women in population-based screening by miRNA expression analysis in cervical scrapes; a feasibility study | Babion et al. [16], 2018, the Netherlands, case-control study | 74 Normal, 139 CIN II-III, 51 SCC, 20 AD, cervical scrapes and frozen tissue | Median: normal (41), CIN III (35), SCC (51) AD (45) | RT-qPCR, RNU24, Mir-423-3p | Mir-9-5p, Mir-149-5p, Mir-203a-3p, Mir-125b-5p, Mir-15b-5p, Mir-375 | Mir-9-5p, Mir-15b-5p | Mir-149-5p, Mir-203a-3p, Mir-375, Mir-125b-5p | Mir-9-5p, Mir-15b-5p | Mir-149-5p, Mir-203a-3p, Mir-125b-5p, Mir-375 | Mir-15b-5p | Mir-149-5p, Mir-375 | ||||
|
|
|||||||||||||||
| 16. Mir-1266 promotes cell proliferation, migration and invasion in cervical cancer by targeting DAB2IP | Wang et al. [43], 2018, China, cohort study | 100 Normal, 50 LSIL, 50 HSIL, 100 CC, serum and frozen tissue | N/A | RT-qPCR, U6snRNA | Mir-1266 | Mir-1266 | Mir-1266 | Mir-1266 | |||||||
|
|
|||||||||||||||
| 17. miRNA detection in cervical exfoliated cells for missed high-grade lesions in women with LSIL/ CIN1 diagnosis after colposcopy-guided biopsy | Ye et al. [40], 2019, China, cross-sectional study | 150 LSIL, 27 HSIL, exfoliated cells and tissue | Mean: LSIL (39.4±6.6), HSIL (39.8±7.1) | RT-qPCR, U6snRNA | miRNA-195, miRNA-29a, miRNA-16– 2, miRNA-20a | miR-NA-16–2, miRNA-20a | miR-NA-195, miRNA-29a | ||||||||
|
|
|||||||||||||||
| 18. Increased expression of Mir-15b is associated with clinicopathological features and poor prognosis in cervical carcinoma | Wen et al. [46], 2017, China, cohort Study | 150 Normal, 124 CIN I, 148 CIN II-III, 185 CC, frozen tissue | Mean: normal (49.23±7.68), CIN I -III (48.60±4.2), CC (49.07±8.52) | RT-qPCR, U6snRNA | Mir-15b | Mir-15b | Mir-15b | Mir-15b | Mir-15b | Mir-15b | |||||
|
|
|||||||||||||||
| 19. Reduced Mir-34a expression in normal cervical tissues and cervical lesions with high-risk human papillomavirus infection | Li et al. [55], 2010, China, case-control study | 64 Normal, 44 CIN I -III, 32 CC, fresh tissue | N/A | RT-PCR and gel electrophoresis, β-actin | Pri-Mir-34a | Pri-Mir-34a | Pri-Mir-34a | Pri-Mir-34a | |||||||
|
|
|||||||||||||||
| 20. Mir-196a targets netrin 4 and regulates cell proliferation and migration of cervical cancer cells | Zhang et al. [41], 2013, China, cross-sectional study | 24 Normal, 13 CIN I, 18 CIN II-III, 15 SCC, frozen tissue | N/A | RT-qPCR, U6snRNA | Mir-196a | Mir-196a | Mir-196a | Mir-196a | |||||||
|
|
|||||||||||||||
| 21. Mir-3156-3p is downregulated in HPV-positive cervical cancer and performs as a tumour-suppressive miRNA | Xie et al. [33], 2017, China, case-control study | 40 Normal, 50 CC, frozen tissue | N/A | RT-qPCR, U6snRNA | Mir-3156-3p | Mir-3156-3p | |||||||||
|
|
|||||||||||||||
| 22. The role of mir-409-3p in regulation of HPV16/18-E6 mRNA in human cervical high-grade squamous intraepithelial lesions | Sommerova et al. [32], 2019, Czech republic, case-control study | 54 Normal, 90 HSIL, FFPE tissue | Median: 36 | RT-qPCR, U6snRNA | Mir-10a-5p, Mir-132-3p, Mir-141-5p, Mir-10b-5p, Mir-34c-5p, Mir-409-3p, Mir-411-5p | Mir-10a-5p, Mir-132-3p, Mir-141-5p | Mir-10b-5p, Mir-34c-5p, Mir-409-3p, Mir-411-5p | ||||||||
|
|
|||||||||||||||
| 23. MicroRNA expression variability in human cervical tissues | Pereira et al. [21], 2010, Portugal, cross-sectional study | 19 Normal, 9 LSIL, 7 HSIL, 4 SCC, frozen tissue | Range: 21-51 | Microarray analysis and validated By RT-qPCR, RNU6B | Mir-26a, Mir-143, Mir-145, Mir-99a, Mir-203, Mir-513, Mir-29a, Mir-199a, Mir-106a, Mir-205, Mir-197, Mir-16, Mir-27a, Mir-142-5p, Mir-148a, Mir-302b, Mir-10a, Mir-196a, Mir-132, Mir-522, Mir-512-3p | Mir-148a, Mir-302b, Mir-10a, Mir-196a, Mir-132 | Mir-26a, Mir-143, Mir-145, Mir-99a, Mir-203, Mir-513, Mir-29a, Mir-199a | Mir-522 and Mir-512-3p, Mir-148a, Mir-302b, Mir-10a, Mir-196a, Mir-132 | Mir-26a, Mir-143, Mir-145, Mir-99a, Mir-203, Mir-513, Mir-29a, Mir-199a, Mir-106a, Mir-205, Mir-197, Mir-16, Mir-27a, Mir-142-5p | Mir-106a, Mir-205, Mir-197, Mir-16, Mir-27a and Mir-142-5p, Mir-148a, Mir-302b, Mir-10a, Mir-196a, Mir-132 | Mir-26a, Mir-143, Mir-145, Mir-99a, Mir-203, Mir-513, Mir-29a, Mir-199a, Mir-522, Mir-512-3p | ||||
|
|
|||||||||||||||
| 24. Expression of Mir200a, Mir93, metastasis-related gene RECK and MMP2/ MMP9 in human cervical carcinoma-relationship with prognosis | Wang et al. [50], 2013, China, cohort study | 100 Normal, 99 SCC, 17 AD frozen tissue | Mean: 49.3±2.39 | RT-qPCR, U6snRNA | Mir93, Mir200a | Mir93, Mir200a | |||||||||
|
|
|||||||||||||||
| 25. Alterations in microRNAs Mir-21 and let-7a correlate with aberrant STAT3 signaling and downstream effects during cervical carcinogenesis | Shishodia et al. [24], 2015, India, cross-sectional study | 23 Normal, 10 LSIL, 13 HSIL, 56 CC, fresh tissue | N/A | RT-qPCR, U6snRNA | Mir-21, Let-7a | Mir-21 | Let-7a | Mir-21, Let-7a | Mir-21 | Mir-21 | Let-7a | ||||
|
|
|||||||||||||||
| 26. Interferon-β induced microRNA--129-5p down-regulates HPV-18 E6 and E7 viral gene expression by targeting SP1 in cervical cancer cells | Wang et al. [43], 2019, China, cross-sectional study | 4 Normal, 6 CIN I-II, 13 CIN III, 14 CC, frozen tissue | Range: 28-77 | Microarray analysis and validated by RT-qPCR, RNU6B | Mir-129-5p | Mir-129-5p | Mir-129-5p | Mir-129-5p | Mir-129-5p | Mir-129-5p | |||||
|
|
|||||||||||||||
| 27. Dysregulated microRNAs involved in the progression of cervical neoplasm | Zeng et al. [51], 2015, China, cross-sectional study | 16 Normal, 18 LSIL, 38 HSIL, 43 SCC, FPPE tissue | Mean: Normal (46.65±4.46), LSIL (42.80±10.03), HSIL (45.65±10.22), SCC (50.08±8.47) | Microarray analysis and validated by RT-qPCR, U6snRNA | Mir-21, Mir-218, Mir-376a, Mir-31, Mir-9, Mir-195, Mir-497, Mir-199b-5p | Mir-21, Mir-31, Mir-9 | Mir-376a, Mir-218, Mir-195, Mir-497, Mir-199b-5p | Mir-218, Mir-195, Mir-497, Mir-199b-5p | |||||||
|
|
|||||||||||||||
| 28. Altered microRNA expression associated with chromosomal changes contributes to cervical carcinogenesis | Wilting et al. [19], 2013, the Netherlands, cross-sectional study | 10 Normal, 18 CIN II-III, 10 SCC, 9 AD, frozen tissue | Range: 30-72 | Microarray analysis and validated by RT-qPCR, RNU43 | Mir-9, Mir-15b, Mir-28-5p, Mir-21, Mir-100, Mir-125b, Mir-375, Mir-203 | Mir-9, Mir-15b, Mir-28-5p, Mir-21 | Mir-100, Mir-125b, Mir-375, Mir-203 | Mir-28-5p, Mir-21 | Mir-203 | Mir-9, Mir-15b, Mir-28-5p, Mir-21 | Mir-100, Mir-125b, Mir-375, Mir-203 | ||||
|
|
|||||||||||||||
| 29. Let-7c is a candidate biomarker for cervical intraepithelial lesions: a pilot study | Malta et al. [22], 2015, Portugal, case-control study | 38 Normal, 14 LSIL, 21 HSIL, exfoliated cells | Mean: 40±13.0 | RT-qPCR, Mir-23a | Let-7c | Let-7c | |||||||||
|
|
|||||||||||||||
| 30. Folate inhibits Mir-27a-3p expression during cervical carcinoma progression and oncogenic activity in human cervical cancer cells | Wang et al. [49], 2020, China, case-control study | 30 Normal, 30 HSIL, 40 SCC, frozen tissue | Mean: sufficient folate group (47.40±2.41), insufficient folate group (48.40±1.82) | Microarray analysis and validated by RT-qPCR, U6snRNA | Mir-27a-3p | Mir-27a-3p | Mir-27a-3p | Mir-27a-3p | |||||||
|
|
|||||||||||||||
| 31. MicroRNAs are biomarkers of oncogenic human papillomavirus infections | Wang et al. [48], 2014, China, case-control study | 38 Normal, 13 CIN I-II, 39 CIN III, 68 CC, FFPE tissue | N/A | Microarray analysis and validated by RT-qPCR, U6snRNA | Mir-378, Mir-27a | Mir-378, Mir-27a | Mir-378, Mir-27a | Mir-27a | |||||||
|
|
|||||||||||||||
| 32. Evaluation of microRNA-205 expression as a potential triage marker for patients with low-grade squamous intraepithelial lesions | Xie et al. [33], 2017, Sweden, cross-sectional study | 16 Normal, 29 CIN I, 44 CIN II, 47 CIN III, 4 CC, exfoliated cells | Median: 32.5 | RT-qPCR, RNU6B | No significance | ||||||||||
|
|
|||||||||||||||
| 33. Complementarity between miRNA expression analysis and DNA methylation analysis in hr-HPV positive cervical scrapes for the detection of cervical disease | Babion et al. [17], 2019, The Netherlands, case-control study | 52 Normal, 25 CIN I, 48 CIN II, 55 CIN III, 24 SCC, 5 AD, exfoliated cells | Median: normal (39.5), CIN I (35.0), CIN II (34.5), CIN3 (35.0), SCC (48.5), AD (50.0) | RT-qPCR, RNU24, Hsa-Mir-423-3p | Mir-15b, Mir-125b, Mir-149, Mir-203a, Mir-375, Let-7b, Mir-93, Mir-222 | Mir-15b, Mir-125b, Mir-149, Mir-203a, Mir-375, Let-7b, Mir-93, Mir-222 | |||||||||
|
|
|||||||||||||||
| 34. Methylation and expression of miRNAs in precancerous lesions and cervical cancer with HPV16 infection | Jiménez-Wences et al. [25], 2016, Mexico, cross-sectional study | 17 Normal, 16 LSIL, 11 CC, exfoliated cells and fresh tissue | Mean: normal HPV-(32.2), normal HPV+ (29.9), LSIL (29.9), CC (52.8) | RT-qPCR, Mir-92a | Mir-124, Mir-218, Mir-193b | Mir-193b | Mir-124, Mir-218 | ||||||||
|
|
|||||||||||||||
| 35. Mir-23b as a potential tumor suppressor and Its regulation by DNA methylation in cervical cancer | Campos-Viguri et al. [26], 2015, Mexico, cross-sectional study | 18 Normal, 19 LSIL, 7 HSIL, 28 CC, exfoliated cells and fresh tissue | N/A | RT-qPCR, Mir-92a | Mir-23b | Mir-23b | Mir-23b | ||||||||
|
|
|||||||||||||||
| 36. Methylation-mediated silencing and tumour suppressive function of hsa-mir-124 in cervical cancer | Wilting et al. [18], 2010, the Nether-lands, cross-sectional study | 23 Normal, 36 CIN I, 48 CIN II-III, 38 SCC and 20 AD, exfoliated cells, frozen tissue and FFPE tissue | Mean: 40.3 | RT-qPCR, RNU43 | Mir-124 | Mir-124 | Mir-124 | ||||||||
|
|
|||||||||||||||
| 37. A correlational study on Mir-34s and cervical lesions | Liu et al. [39], 2018, China, case-control study | 30 Normal, 30 CIN1, 30 CIN2/3, 30 CC, frozen tissue | Mean: 45.77±10.73 | RT-qPCR, U6snRNA | miRNA-34 | miRNA-34 | |||||||||
|
|
|||||||||||||||
| 38. Clinical value of combined detection of Mir-1202 and Mir-195 in early diagnosis of cervical cancer | Yang et al. [45] 2019, China, case-control study | Mean: normal (41.1±2.8), CC (40.2±3.7) | RT-qPCR, U6snRNA | Mir-1202, Mir-195 | Mir-1202, Mir-195 | ||||||||||
CC, cervical cancer; CIN, cervical intraepithelial neoplasia; LSIL, low-grade squamous intraepithelial lesion; HSIL, high-grade squamous intraepithelial lesion; NILM, negative for intraepithelial lesion or malignancy; FFPE, formalin-fixed paraffin embedded; RT-qPCR, quantitative reverse transcription polymerase chain reaction; PDK, pyruvate dehydrogenase kinase; N/ A, not available; ASCUS, atypical squamous cells of undetermined significance; SCC, squamous cell carcinoma; HPV, human papillomavirus; AD, adenocarcinoma; ICC, intrahepatic cholangiocarcinoma; PAP, poly A polymerase; DAB2IP, disabled homolog 2-interacting protein; RECK, reversion-inducing-cysteine-rich protein; MMP, matrix metalloproteinases.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download