Abstract
Background
Zika virus (ZIKV) is a major health concern worldwide, highlighting the importance of accurate and reliable viral detection to control transmission. The careGENE™ ZIKV Reverse Transcription-Polymerase Chain Reaction (RT-PCR) Kit (Wells Bio, INC, Korea) was developed to detect Zika infection. We compared the clinical sensitivity and specificity of this kit with those of the Aptima Zika Virus Assay (Hologic, Inc., USA) and the World Health Organization (WHO)-recommended conventional PCR method.
Methods
A total of 168 urine samples (143 clinical samples and 25 RNA-spiked samples [RNA-positive lyophilized plasma spiked in urine]) were tested in this study.
Results
In comparison with the WHO-recommended conventional PCR method, the careGENE™ ZIKV RT-PCR Kit detected ZIKV RNA with a sensitivity of 100% (95% confidence interval [CI], 73.5–99.9) and specificity of 100% (95% CI, 88.9–97.8). Similar results were obtained with the Aptima Zika Virus Assay, as evident from a sensitivity of 100% (95% CI, 73.5–99.9) and specificity of 100% (95% CI, 88.9–97.8). The limit of detection at a 95% detection probability was 1 U/mL in the urine.
초록
배경
지카바이러스는 모기 매개 감염원이며 국제 공중보건 위기관리 대상으로, 바이러스의 확산을 방지하기 위해서는 지카바이러스를 정확하게 검출하는 것이 중요하다. 이에 본 연구에서는 지카바이러스를 검출하기 위해 고안된 역전사 중합효소 연쇄반응 키트인 CareGENE™ Zika virus RT-PCR kit (Wells Bio, Inc., Korea)의 민감도와 특이도를 Aptima Zika Virus Assay (Hologic, Inc., USA)와 WHO 추천 검사인 PCR 검사로 비교 평가하고자 하였다.
Zika virus (ZIKV), a member of the Flaviviridae family, was first isolated in 1947 from the serum of a febrile rhesus monkey in Uganda [1]. ZIKV is mostly transmitted to humans via mosquitoes, mainly Aedes aegypti and less commonly by A. albopictus. In 2007, a ZIKV outbreak in humans occurred on the island of Yap, Micronesia, and spread throughout Oceania until late 2013 [2]. In May 2015, ZIKV infection was confirmed in Brazil and continued to spread to the continent of South America [3]. The World Health Organization (WHO) classified ZIKV as a public health emergency of international concern, and recent data reported ZIKV transmission in about 75 countries [4]. Traveling has contributed to the expansion of ZIKV to non-endemic countries. For example, the first ZIKV infection case in Korea in 2016 was reported in a traveler who had returned from Brazil [5]. Since then, ZIKV infections have been reported in Korea in travelers from south-eastern Asia and South Africa [6].
ZIKV can be transmitted not only through vector-borne routes but also through non-vector-borne routes such as sexual intercourse, transplacental transmission, blood product transfusion, and body fiuids. More than 70% of ZIKV-infected patients are asymptomatic [7]. Even in symptomatic patients, the clinical conditions such as maculopapular rash, mild fever, arthralgia, conjunctivitis, myalgia, and headache are similar to the symptoms of dengue virus or other mosquito-borne fiavivirus-related infections, and can be confused with other causes of febrile illness [8]. The most significant consideration in ZIKV-infected patients is the causal relationship between ZIKV infection in pregnant women and the development of birth defects [9]. Therefore, prevention of ZIKV infection in pregnant women is imperative.
ZIKV infection can be diagnosed through the detection of ZIKV-specific immunoglobulin M (IgM) antibodies and from the presence of ZIKV RNA or antigens. According to the WHO guidance, IgM antibody positivity alone cannot be deemed sufficient for the diagnosis of ZIKV infections [10]. Instead, ZIKV infection is characterized with the presence of ZIKV RNA or antigens in the serum or other samples, IgM antibodies against ZIKV, or anti-Zika antibodies in the plaque reduction neutralization test. As plaque reduction neutralization test is labor-intensive [10], the diagnosis of ZIKV infection mainly relies on the detection of ZIKV RNA. ZIKV RNA detection using reverse transcription-PCR (RT-PCR) is useful during the acute phase, which encompasses the first 7 days from symptom onset. RT-PCR-mediated detection of ZIKV using urine, serum, and saliva samples proved urine samples to be the most reliable and sensitive for identification of acute ZIKV infection [11, 12].
Here, we evaluated the analytical performance of a newly developed real-time RT-PCR kit (careGENE™ Zika virus RT-PCR Kit; Wells Bio. Inc., Seoul, Korea) to detect ZIKV RNA in urine samples.
A total of 168 urine specimens were analyzed in this study (Fig. 1). In total, 127 clinical specimens of symptomatic patients were provided by Boca Biolistics (Pompano Beach, FL, USA); these specimens were collected from patients with ZIKV-like symptoms. Clinical information on the samples from Boca Biolistics LLC is given in Table 1. In addition, 25 ZIKV-positive samples were sourced from the dilution of ZIKV RNA-containing lyophilized plasma, which is the candidate WHO international standard for ZIKV RNA nucleic acid amplification technique-based assays. In addition, 16 negative samples from asymptomatic pregnant Korean women were obtained. The clinical sample collection protocols were approved by the Bioethics Committee of the Hospital de la plaza de la Salud (BB-ID-061; Santo Domingo, Dominican Republic) and the Institutional Review Board of Korea University Guro Hospital (MD16075; Seoul, Korea).
Twenty-five natural positive urine specimens were provided by Boca Biolistics, and were confirmed positive for ZIKV RNA using the Aptima Zika Virus Assay (Hologic, Inc., San Diego, CA, USA). Twenty-five positive specimens were prepared by spiking the negative urine samples with ZIKV RNA. Positive RNA was the candidate for the WHO international standard (PEI code 11468/16; Paul-Ehrlich-Institute, Langen, Germany) and included 0.5 mL of lyophilized plasma and the non-infectious Asian strain of ZIKV RNA containing 50,000,000 U/mL. This WHO standard ZIKV was diluted by negative urine specimens obtained from Boca Biolistics that were confirmed as ZIKV negative using the Aptima Zika Virus Assay. The spiked samples were prepared at concentrations of 1, 2, 3, and 4 U/mL of ZIKV RNA.
A total of 118 negative urine specimens were included in this study, of which 102 were provided by Boca Biolistics; 85 negative urine specimens were obtained from non-pregnant individuals, and 17, from pregnant women. All specimens were confirmed as ZIKV negative using the Aptima Zika Virus Assay. In addition, 16 negative urine specimens were collected from asymptomatic pregnant women who visited Korea University Guro Hospital.
The QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) was used for RNA extraction. Urine (140 μL) sample was added to a 1.5-mL microcentrifuge tube containing 560 μL of AVL buffer with carrier RNA. The mixture was incubated at room temperature (20-22°C) for 10 minutes. Ethanol (560 μL) was added to the mixture and mixed by pulse-vortexing for 15 seconds. After mixing, the tube was briefiy centrifuged to collect droplets from the inside of the lid and sides to the bottom. The resulting solution (630 μL) was carefully applied to the QIAamp Mini column equipped with a 2-mL collection tube without a wetting rim. The cap of the column was closed and the column was centrifuged at 6,000×g for 1 minute. To wash the column, the cap was opened and 500 μL of AW1 buffer was added. The column was centrifuged at 6,000×g for 1 minute. The column was washed again with 500 μL of AW2 buffer, and the RNA was eluted with 60 μL of nuclease-free water.
Real-time PCR was performed to amplify NS5B and envelope genes of ZIKV using careGENE™ ZIKV RT-PCR Kit (Wells Bio. Inc.) on an Applied Biosystems 7500 Real-Time PCR system (Life Technologies, Carlsbad, CA, USA). The assay was performed in a 15 μL reaction mixture containing 5 μL of 4×1 Step RT-PCR Mix, 5 μL of Zika primer/probe mix, and 5 μL of nuclease-free water. The mixture components were prepared on ice, and 5 μL of extracted RNA was well mixed with 15 μL of mixture components. The optimized thermal cycling conditions were as follows: reverse transcription at 50°C for 15 minutes, initial denaturation at 95°C for 20 seconds, and 40 cycles of PCR amplification at 95°C for 15 sec and 58°C for 60 seconds. Probes for ZIKV-specific sequences were labeled with the fiuorophores FAM and CY5/Alexa647 for dual detection. The probe for the internal control (IC)-specific sequence was labeled with the fiuorophore VIC or HEX. The careGENE™ ZIKV RT-PCR Kit includes an endogenous IC, which can be used to control the sample preparation procedure (nucleic acid extraction) and/or as an RT-PCR inhibition control (Table 2). The real-time PCR results were defined based on the following criteria: the sample was positive if the Ct value was <36 with acceptable results or considered negative if the Ct value was ≥36 (Table 3). Representative results of positive and negative samples were shown on Fig. 2.
To test sensitivity, the WHO standard ZIKV was diluted with negative urine specimens to target concentrations of 1, 2, 3, and 4 U/mL of ZIKV RNA; 1 U/mL of ZIKV RNA-containing urine was tested 10 times. To evaluate the specificity in pregnant women, 17 ZIKV-negative urine specimens, which were confirmed as ZIKV-negative using the Aptima Zika Virus Assay, were collected from symptomatic pregnant women and 16 samples were confirmed as ZIKV-negative from non-symptomatic pregnant women using conventional PCR. Additionally, 85 negative urine samples from non-pregnant symptomatic individuals confirmed as ZIKV negative using the Aptima Zika Virus Assay were analyzed.
To confirm all positive and negative results, conventional PCR was performed. Amplification was carried out using the ZIKV primer pairs targeting NS4A (Table 4). PCR was carried out using an initial activation step at 94°C for 3 minutes, followed by 30 cycles of 94°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds, with a final extension step for 7 minutes at 72°C. Conventional PCR products were detected by agarose gel electrophoresis, and positivity of amplification products was confirmed following comparison with positive control bands.
Comparisons of the methods were made using the chi-square agreement analysis and the kappa value. All statistical analyses were performed using MedCalc Statistical Software version 18.5 (MedCalc Software bvba, Ostend, Belgium; http://www.medcalc.org; 2018).
Twenty-five naturally positive specimens that were previously confirmed to be positive using the Aptima Zika Virus Assay and 25 positive RNA-spiked specimens from the Paul-Ehrlich-Institute were tested positive for ZIKV using the careGENE™ ZIKV RT-PCR Kit and the conventional PCR method (Table 5). For the clinical sensitivity evaluation, 25 ZIKV-infected clinical urine specimens tested positive with 100% sensitivity. These clinical specimens were obtained between 2 and 10 days after the onset of symptoms; 24 of 25 specimens were obtained within 7 days of symptom onset (Table 1). Lyophilized ZIKV RNA-containing samples were diluted with negative urine samples to concentrations of 1, 2, 3, and 4 U/mL of ZIKV RNA. The detection limit of the assay was 1 U/mL for ZIKV RNA; 13 repeat measurements of 1 U/mL of ZIKV RNA-containing samples tested positive. Ct values of RNA-spiked samples were detected by real-time PCR and found to be between 28 and 32 for NS2B gene and between 32 and 36 for E gene.
For the specificity evaluation, 85 negative specimens from non-pregnant individuals and 33 negative specimens from pregnant women were tested using the careGENE™ ZIKV RT-PCR Kit and the conventional PCR method, and all samples were deemed negative. Of these samples, 102 also tested negative using the Aptima Zika Virus Assay. In particular, 16 urine specimens from asymptomatic pregnant women were included and tested negative, indicating 100% specificity.
An agreement evaluation was performed between the Aptima Zika Virus Assay and careGENE™ ZIKV RT-PCR Kit (N=127) and between the conventional PCR method and careGENE™ Zika Virus RT-PCR Kit (N=168). The positive and negative percent agreement values between the careGENE™ Kit and Aptima assay were both 100%. The kappa value was 1.00 and the chi-squared value was 126.0 (Table 6). Further, the positive and negative percent agreement values between the careGENE™ Kit and conventional PCR were both 100%. The kappa value was 1.00 and the chi-squared value was 167.0. The diagnostic sensitivity and specificity were both 100%.
The clinical signs and symptoms of ZIKV infection are similar to those of dengue virus or other mosquito-borne fiavivirus-related infections. The most common signs and symptoms include maculopapular rash, mild fever, arthralgia, conjunctivitis, myalgia, and headache [8]. However, more than 70% of infections are asymptomatic [7]. The most significant clinical condition is neonatal infection. ZIKV-associated congenital defects owing to vertical viral transmission from the mother to the fetus are clinically significant and have been well documented [9]. Aside from mosquito bites, ZIKV can be transmitted through sexual contact. Further, ZIKV infection via transfusion of infected blood products remains a significant concern. About 1.1% and 2.8% of asymptomatic blood donors were tested positive for ZIKV during their respective outbreaks [11, 13]. Moreover, immunocompromised patients can be infected by transfusion from an asymptomatic ZIKV-infected blood donor [7]. ZIKV infection through blood transfusion to immunocompromised patients such as transplant recipients has been recently reported [7, 8]. Therefore, ZIKV screening of asymptomatic individuals such as pregnant women, travelers, and blood donors should be emphasized. For the ZIKV screening test, a sensitive detection method is warranted.
There is no gold standard for the diagnosis of ZIKV infection, but detection of ZIKV-specific IgM antibodies and presence of ZIKV RNA are the commonly employed methods. For the diagnosis of ZIKV infection, antibody detection methods such as enzyme-linked immunosorbent assays have shown relatively lower sensitivity and specificity than ZIKV RNA detection methods such as real-time PCR [14]. Therefore, ZIKV infection is easily confirmed by real-time PCR detection of ZIKV RNA.
The ZIKV genome is composed of three structural units, including the capsid, membrane precursor/membrane, and envelope proteins, and seven non-structural proteins. There are various molecular assays to detect ZIKV, but only a few were developed to detect two gene targets [15]. The careGENE™ ZIKV RT-PCR Kit was developed based on real-time RT-PCR technology exploiting an RT reaction to convert RNA into complementary DNA, followed by PCR to amplify the specific target sequences (E and NS5B genes) and target-specific probes for detection of the amplified DNA. ZIKV genetic variations have been reported as two lineages (African and Asian) and three genotypes (West African, East African, and Asian). Up to 10 nucleotide mismatches have been identified between a previously published assay and Asian lineage consensus sequences [16]. Therefore, a ZIKV RNA detection test designed to detect a single RNA target may produce false-negative results, whereas multiple target ZIKV RNA detection can reduce false-negative results. The careGENE™ ZIKV RT-PCR Kit using dual detection of the E and NS5B genes of ZIKV with FAM and CY5/Alexa647 probes could increase the sensitivity and specificity of ZIKV detection.
In this study, 1 U/mL of ZIKV RNA was successfully detected using the careGENE™ ZIKV RT-PCR Kit. The analytical specificity of the careGENE™ kit was 100% as compared with a previously approved kit. The sensitivity results showed maximum sensitivity as compared with the previously reported limit of detection concentration. Thus, the careGENE™ ZIKV RT-PCR Kit may be useful for the screening of ZIKV RNA and diagnosis of ZIKV infection.
ZIKV RNA can be detected at higher levels and for a longer time after the onset of infection in the urine than in the serum [11, 12, 17]. ZIKV RNA can be detected in the urine from 2 to more than 20 days following symptom onset. However, RT-PCR for both serum and urine samples is recommended for diagnostic molecular testing within 7 days from symptom onset [18]. ZIKV RNA is detectable in the urine from the first day of symptom onset to day 20 [11]. ZIKV RNA was less detectable in the serum than in the urine from the first week after symptom onset. Therefore, urine samples are preferred over serum samples, especially within 1 week of symptom onset. In our study, 24 of 25 clinical ZIKV specimens were obtained from patients within 7 days of symptom onset (Table 1). Our results show that the appropriate sensitivity of urine RT-PCR testing was within 7 days. The detection limit and repeatability of the assay were 1 IU/mL and 100%, respectively, for the urine specimens.
In this study, a single-blind, single-center clinical test was performed to evaluate the ability of the careGENE™ ZIKV RT-PCR Kit to qualitatively detect ZIKV RNA from human urine specimens and its clinical sensitivity and specificity were assessed. In addition, the performance of the kit was compared to that of the WHO-recommended conventional PCR method. The careGENE™ ZIKV RT-PCR Kit was used to detect Zika infection from 168 human urine samples (50 positive and 118 negative). Positive identification of Zika infection was selective and specific for the target Zika viral sequence. Overall, the performance of the careGENE™ ZIKV RT-PCR Kit was satisfactory. Both the sensitivity and specificity were 100%.
The careGENE™ ZIKV RT-PCR Kit is a sensitive assay to detect ZIKV RNA in urine specimens. The detection limit of the assay was 1 U/mL in the urine, which is deemed satisfactory for a diagnostic test.
Acknowledgements
This study was supported by a government-wide R&D fund project for infectious disease research (HG18C0012), National Research Foundation of Korea (NRF-2016R1A5A1010148) and a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HR20C0021).
REFERENCES
1. Dick GW, Kitchen SF, Haddow AJ. 1952; Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg. 46:509–20. DOI: 10.1016/0035-9203(52)90042-4. PMID: 12995440.

2. Waggoner JJ, Pinsky BA. 2016; Zika virus: diagnostics for an emerging pandemic threat. J Clin Microbiol. 54:860–7. DOI: 10.1128/JCM.00279-16. PMID: 26888897. PMCID: PMC4809954.

3. Campos GS, Bandeira AC, Sardi SI. 2015; Zika virus outbreak, Bahia, Brazil. Emerg Infect Dis. 21:1885–6. DOI: 10.3201/eid2110.150847. PMID: 26401719. PMCID: PMC4593454.

4. Jimenez A, Shaz BH, Kessler D, Bloch EM. 2017; How do we manage blood donors and recipients after a positive Zika screening result? Transfusion. 57:2077–83. DOI: 10.1111/trf.14252. PMID: 28734023.

5. Jang HC, Park WB, Kim UJ, Chun JY, Choi SJ, Choe PG, et al. 2016; First imported case of Zika virus infection into Korea. J Korean Med Sci. 31:1173–7. DOI: 10.3346/jkms.2016.31.7.1173. PMID: 27366020. PMCID: PMC4901014.

6. Yoon D, Shin SH, Jang HC, Kim ES, Song EH, Moon SM, et al. 2017; Epidemiology and clinical characteristics of Zika virus infections imported into Korea from March to October 2016. J Korean Med Sci. 32:1440–4. DOI: 10.3346/jkms.2017.32.9.1440. PMID: 28776338. PMCID: PMC5546962.

7. Barjas-Castro ML, Angerami RN, Cunha MS, Suzuki A, Nogueira JS, Rocco IM, et al. 2016; Probable transfusion-transmitted Zika virus in Brazil. Transfusion. 56:1684–8. DOI: 10.1111/trf.13681. PMID: 27329551.

8. Darrigo LG Jr. 2018; , de Sant'Anna Carvalho AM, Machado CM. Chikungunya, Dengue, and Zika in immunocompromised hosts. Curr Infect Dis Rep. 20:5. DOI: 10.1007/s11908-018-0612-2. PMID: 29551005. PMCID: PMC5857271.

9. Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. 2016; Zika virus and birth defects--reviewing the evidence for causality. N Engl J Med. 374:1981–7. DOI: 10.1056/NEJMsr1604338. PMID: 27074377.
10. World Health Organization. Zika virus disease: interim case definitions. http://www.who.int/iris/handle/10665/204381. Updated on Feb 2016.
11. Bingham AM, Cone M, Mock V, Heberlein-Larson L, Stanek D, Blackmore C, et al. 2016; Comparison of test results for Zika virus RNA in urine, serum, and saliva specimens from persons with travel-associated Zika virus disease - Florida, 2016. MMWR Morb Mortal Wkly Rep. 65:475–8. DOI: 10.15585/mmwr.mm6518e2. PMID: 27171533.

12. Lamb LE, Bartolone SN, Kutluay SB, Robledo D, Porras A, Plata M, et al. 2016; Advantage of urine based molecular diagnosis of Zika virus. Int Urol Nephrol. 48:1961–6. DOI: 10.1007/s11255-016-1406-9. PMID: 27567913.

13. Musso D, Nhan T, Robin E, Roche C, Bierlaire D, Zisou K, et al. 2014; Potential for Zika virus transmission through blood transfusion demonstrated during an outbreak in French Polynesia, November 2013 to February 2014. Euro Surveill. 19:20761. DOI: 10.2807/1560-7917.ES2014.19.14.20761. PMID: 24739982.

14. Balmaseda A, Zambrana JV, Collado D, García N, Saborío S, Elizondo D, et al. 2018; Comparison of four serological methods and two reverse transcription-PCR assays for diagnosis and surveillance of Zika virus infection. J Clin Microbiol. 56:e01785–17. DOI: 10.1128/JCM.01785-17. PMID: 29305550. PMCID: PMC5824050.

15. Theel ES, Hata DJ. 2018; Diagnostic testing for Zika virus: a postoutbreak update. J Clin Microbiol. 56:e01972–17. DOI: 10.1128/JCM.01972-17. PMID: 29386264. PMCID: PMC5869840.

16. Corman VM, Rasche A, Baronti C, Aldabbagh S, Cadar D, Reusken CB, et al. 2016; Assay optimization for molecular detection of Zika virus. Bull World Health Organ. 94:880–92. DOI: 10.2471/BLT.16.175950. PMID: 27994281. PMCID: PMC5153932.

17. Rossini G, Gaibani P, Vocale C, Cagarelli R, Landini MP. 2017; Comparison of Zika virus (ZIKV) RNA detection in plasma, whole blood and urine - Case series of travel-associated ZIKV infection imported to Italy, 2016. J Infect. 75:242–5. DOI: 10.1016/j.jinf.2017.05.021. PMID: 28648495.
18. Centers for Disease Control and Prevention. Interim guidance for Zika virus testing of urine-United States, 2016. http://dx.doi.org/10.15585/mmwr.mm6518e1. Updated on May 2016. DOI: 10.15585/mmwr.mm6518e1. PMID: 27171368.
Fig. 1
Information on the 168 urine specimens analyzed in this study.
Abbreviations: ZIKV, zika virus; RT-PCR, reverse transcription-PCR.
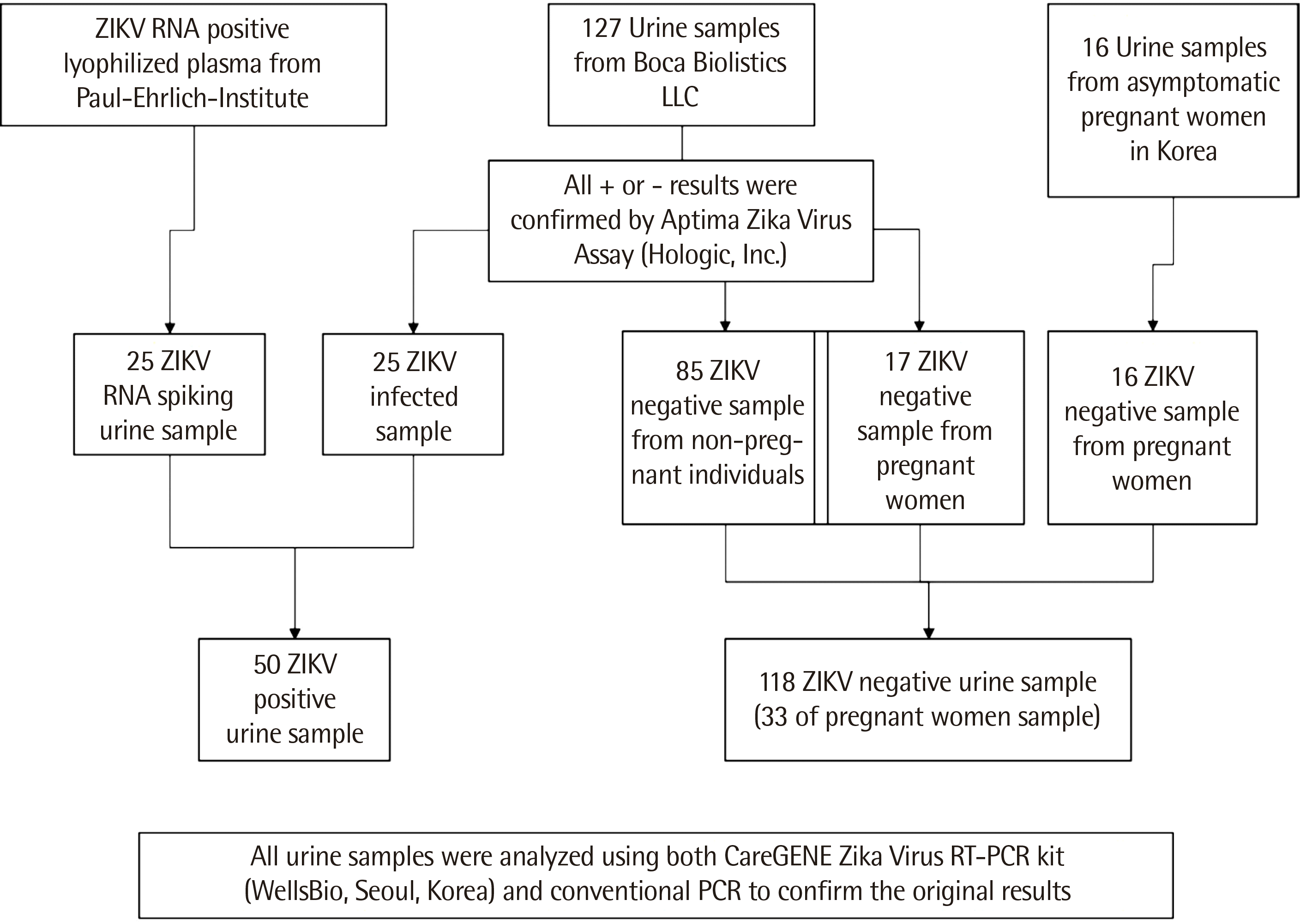
Fig. 2
(A) Representative results of ZIKV-positive sample, both the target gene for ZIKV (blue, green) and the internal control gene (pink) were amplified. (B) Representative results of ZIKV-negative sample, only the internal control gene (pink) was amplified.
Abbreviations: ZIKV, zika virus; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
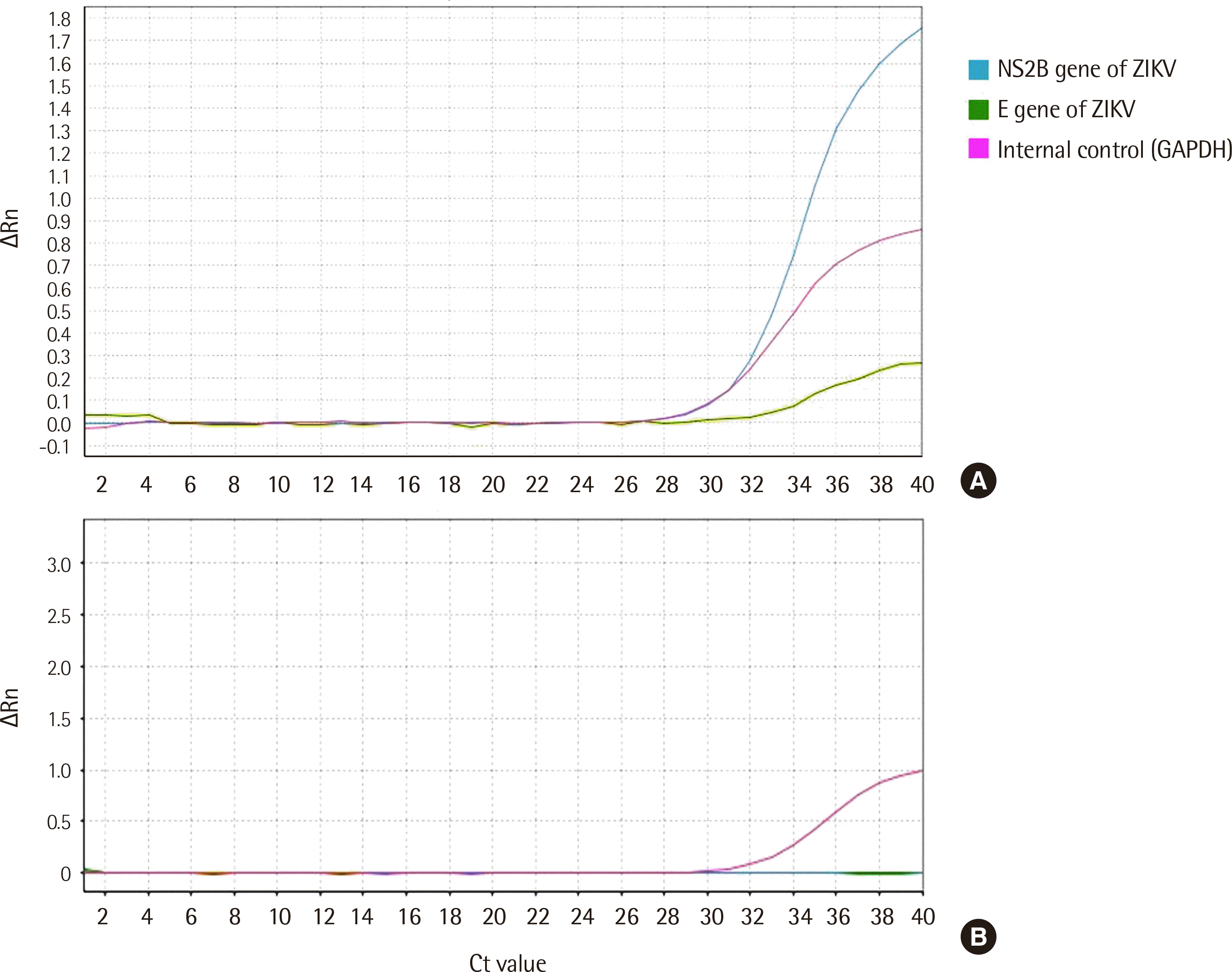
Table 1
Clinical information on the clinical samples from naturally ZIKV-infected or uninfected patients with ZIKV symptoms provided by Boca Biolistics LLC (N=127)
| Symptom onset date | N of patients with ZIKV infection confirmed by ZIKV PCR |
N of non-infectious patients* N of non-pregnant subjects (N of pregnant subjects)* |
|---|---|---|
| 1 | 1 | |
| 2 | 1 | 14 (1) |
| 3 | 3 | 26 (3) |
| 4 | 8 | 22 (2) |
| 5 | 7 | 22 (2) |
| 6 | 4 | 5 (1) |
| 7 | 1 | 2 (1) |
| 8 | 3 (2) | |
| 9 | 1 | |
| ≥10 | 1† | 6‡ (5) |
| Total | 25 | 102 (17) |
Table 2
Target gene and reporter dye composition of the careGENE™ ZIKV RT-PCR Kit
| Detection target | Target gene | Reporter |
|---|---|---|
| Zika virus RNA target 1 | NS2B | FAM |
| Zika virus RNA target 2 | E | CY5/Alexa647 |
| Endogenous internal control (enIC) | GAPDH | VIC/HEX |
Table 3
Acceptance criteria for positive results from the careGENE™ ZIKV RT-PCR Kit
| Ct | Zika-FAM | Zika-Cy5/Alexa647 | enIC | Result |
|---|---|---|---|---|
| <36 | + | + | + | Positive |
| + | - | + | Weak positive | |
| - | + | + | Weak positive | |
| ~36 | +/- | - | + | Negative |
| - | +/- | + | Negative | |
| >36 | - | - | + | Negative |
| - | - | - | - | Invalid |
Table 4
Forward and reverse primer sets for the conventional PCR amplification of ZIKV
| Primer | Sequence | MW | TM (°C) | Size (bp) |
|---|---|---|---|---|
| Forward | GCAGAGCAATGGATGGGA | 5,941.46 | 58 | 97 |
| Reverse | CTGAGGGCATGTGCAAACC | 5,779.46 | 58.5 |
Table 5
Clinical evaluation results of the careGENE™ ZIKV RT-PCR Kit
Table 6
Contingency table for the new test method (careGENE) and comparative methods (Aptima assay and conventional PCR)




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download