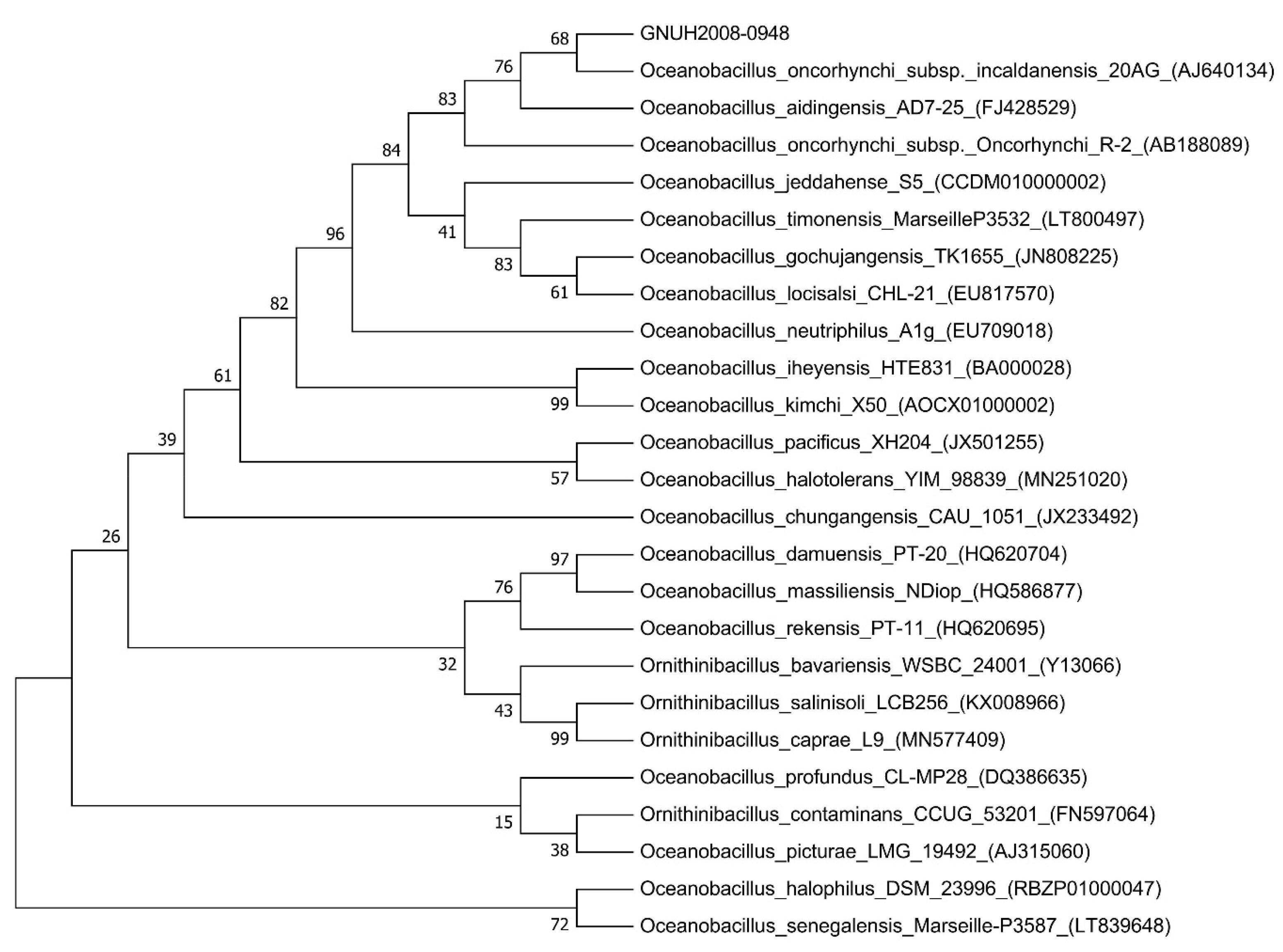
 | Fig. 1Neighbor-joining phylogenetic tree based on 16S rRNA gene sequences of 24 similar species, including the Oceanobacillus oncorhynchi subsp. incaldanensis strain (GNUH2008-0948) isolated from a 69-year-old male patient in South Korea, generated using MEGA7 (Molecular Evolutionary Genetics Analysis version 7.0, Kumar, Stecher, and Tamura 2015). Numbers at each node are percentages at which the associated taxa clustered together in the bootstrap test (1,000 replicates). |
Dear Editor,
The genus Oceanobacillus comprises gram-positive bacilli that were first detected in deep-sea sediments [1]. To date, 36 Oceanobacillus species have been isolated from various sources, including sea fish, spring water, soils, food, and the human gut [2-6]. These bacteria can be easily misinterpreted as contaminants, because they cannot be identified with the typical phenotypic methods used in clinical laboratories. Moreover, their role in human pathogenesis is unknown. We report the first case of human Oceanobacillus oncorhynchi infection.
This study was approved by the Institutional Review Board of Gyeongsang National University Hospital, Jinju, Korea, with a waiver of informed consent (202012007). A 69-year-old male patient was admitted to Gyeongsang National University Hospital in August 2020 because of scrotal pain that had started five days prior to admission. He had no remarkable medical history. He was suspected of having epididymo-orchitis. Laboratory tests showed increased leukocyte counts (11.41×109/L; reference range, 4–10×109/L) and C-reactive protein level (120.8 mg/L; <5 mg/L). Other laboratory blood test results were within the reference ranges. Two sets of aerobic and anaerobic blood culture (BACT/ALERT Culture Media, bioMérieux, Marcy l’Etoile, France) were performed before initiating empiric antimicrobial treatment with ceftriaxone, and gram-positive bacilli were isolated from an aerobic blood culture. The colony morphology on sheep blood agar plate was non-hemolytic, grayish, circular, and smooth, and the cell morphology was thin, straight, and with endospores. We considered this catalase- and oxidase-positive bacillus to be a contaminant. Matrix-associated laser desorption ionization-time of flight mass spectrometry (bioMérieux) and biochemical tests using the VITEK 2 system (bioMérieux) did not provide reliable identification of the bacterium. Therefore, we concluded that the “unidentified gram-positive bacilliform bacterium” was a contaminant. The next day, a closed pus specimen obtained during incision and drainage was submitted for bacterial culture. On Gram staining of a smear, 5–10 leukocytes per high-power field and gram-positive bacilli were observed. The colony morphology was identical to that of the microbes found in the blood cultures. As we recognized that this bacterium could be of clinical significance, we conducted 16S rRNA gene sequencing for species identification. The highest similarity was observed with O. oncorhynchi subsp. incaldanensis (99.6%), followed by O. aidingensis (99.2%) and O. oncorhynchi subsp. oncorhynchi (98.9%), in the EzTaxon database (v2.1; http://www.ezbiocloud.net) (Fig. 1). For a more accurate identification of the isolate, whole-genome sequencing was performed and the data were analyzed using the TrueBac ID-Genome system (www.truebacid.com; ChunLab, Inc., Seoul, Korea) [7]. The bacterium was finally identified as O. oncorhynchi subsp. incaldanensis.
Antimicrobial susceptibility testing using disk diffusion was performed, and the results were interpreted according to the European Committee on Antimicrobial Susceptibility Testing clinical breakpoint table v. 11.0 for Staphylococcus [8]. Inhibition zones for penicillin, ampicillin, erythromycin, clindamycin, and ceftazidime were 28, 24, 13, 12, and 6 mm in diameter, respectively; based on these results, the isolate was considered resistant to these antimicrobials. Inhibition zones for amikacin, gentamicin, tobramycin, and levofloxacin were 36, 36, 38, and 32 mm in diameter, respectively; based on these findings, the isolate was considered susceptible to these antimicrobials. The inhibition zone of 18 mm for ceftriaxone was not interpreted.
Intravenous ceftriaxone (2 g daily) was administered for two days, and the regimen was stepped up to intravenous piperacillin/tazobactam (4.5 g at eight-hour intervals) and tobramycin (80 mg at eight-hour intervals). The wound was kept open with betadine-soaked gauze packing for a week, after which the necrotic tissue was removed. Three days later, the wound improved and was closed. O. oncorhynchi subsp. incaldanensis was not isolated from follow-up cultures. Two weeks later, the medical condition of the patient improved, with no sign of active infection, and the patient was discharged.
Human diseases caused by Oceanobacillus spp. have not been described; however, O. oncorhynchi subsp. incaldanensis can cause infections even in immunocompetent patients. Members of the genus Oceanobacillus are mostly alkaliphilic [2, 3, 9]. It is noteworthy that the bacteremia-causing infection developed in the male reproductive organs, where the pH is relatively alkaline compared with that in other body parts, as an alkaline environment is important for sperm survival [10]. Reportedly, the pH range for growth of O. oncorhynchi subsp. incaldanensis is 6.5–9.5 (optimum pH, 9.0) [9]. This may explain why this bacterium is rarely isolated on sheep blood agar plate. While it does grow on sheep blood agar plate, clinical microbiologists may misinterpret it as a contaminant and therefore not pursue species identification. Because of the underestimation of its clinical significance, current commercial identification systems do not identify Oceanobacillus species. Therefore, infections caused by Oceanobacillus species might be more common than previously thought. Further studies on the pathogenesis, clinical prognosis, virulence factors, and antimicrobial susceptibility of Oceanobacillus species are warranted.
Go to : 
ACKNOWLEDGEMENTS
The pathogen resource for this study was provided by Gyeongsang National University Hospital, a branch of National Culture Collection for Pathogens (GNUH-NCCP, Jinju, Korea). We thank Chang Kun Ha, Young Bin Lee, and Ji Gwang Lee for technical support.
Go to : 
Notes
AUTHOR CONTRIBUTIONS
Conceptualization: Byun JH. Data curation: Kim M, Choi JH, Shin H, Lee WK. Writing-review and editing: Byun JH, Kim M. Final approval of the manuscript: all authors.
RESEARCH FUNDING
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (Ministry of Science and ICT) (NRF-2020R1F1A1066895).
Go to : 
REFERENCES
1. Lu J, Nogi Y, Takami H. 2001; Oceanobacillus iheyensis gen. nov., sp. nov., a deep-sea extremely halotolerant and alkaliphilic species isolated from a depth of 1050 m on the Iheya Ridge. FEMS Microbiol Lett. 205:291–7. DOI: 10.1111/j.1574-6968.2001.tb10963.x. PMID: 11750818.
2. Yumoto I, Hirota K, Nodasaka Y, Nakajima K. 2005; Oceanobacillus oncorhynchi sp. nov., a halotolerant obligate alkaliphile isolated from the skin of a rainbow trout (Oncorhynchus mykiss), and emended description of the genus Oceanobacillus. Int J Syst Evol Microbiol. 55:1521–4. DOI: 10.1099/ijs.0.63483-0. PMID: 16014475.
3. Romano I, Lama L, Nicolaus B, Poli A, Gambacorta A, Giordano A. 2006; Oceanobacillus oncorhynchi subsp. incaldanensis subsp. nov., an alkalitolerant halophile isolated from an algal mat collected from a sulfurous spring in Campania (Italy), and emended description of Oceanobacillus oncorhynchi. Int J Syst Evol Microbiol. 56:805–10. DOI: 10.1099/ijs.0.63575-0. PMID: 16585699.
4. Long X, Ye R, Zhang S, Liu B, Zhang Y, Zeng Z, et al. 2015; Oceanobacillus damuensis sp. nov. and Oceanobacillus rekensis sp. nov., isolated from saline alkali soil samples. Antonie van Leeuwenhoek. 108:731–9. DOI: 10.1007/s10482-015-0529-9. PMID: 26168852.
5. Whon TW, Jung MJ, Roh SW, Nam YD, Park EJ, Shin KS, et al. 2010; Oceanobacillus kimchii sp. nov. isolated from a traditional Korean fermented food. J Microbiol. 48:862–6. DOI: 10.1007/s12275-010-0214-7. PMID: 21221947.
6. Lagier JC, Khelaifia S, Azhar EI, Croce O, Bibi F, Jiman-Fatani AA, et al. 2015; Genome sequence of Oceanobacillus picturae strain S1, an halophilic bacterium first isolated in human gut. Stand Genomic Sci. 10:91. DOI: 10.1186/s40793-015-0081-2. PMID: 26523201. PMCID: PMC4627390.

7. Ha SM, Kim CK, Roh J, Byun JH, Yang SJ, Choi SB, et al. 2019; Application of the whole genome-based bacterial identification system, TrueBac ID, using clinical isolates that were not identified with three matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) systems. Ann Lab Med. 39:530–6. DOI: 10.3343/alm.2019.39.6.530. PMID: 31240880. PMCID: PMC6660342.

8. The European Committee on Antimicrobial Susceptibility Testing. 2021. Breakpoint tables for interpretation of MICs and zone diameters, v. 11.0. https://eucast.org/clinical_breakpoints/. Updated on Jan 2021.
9. Liu W, Yang SS. 2014; Oceanobacillus aidingensis sp. nov., a moderately halophilic bacterium. Antonie van Leeuwenhoek. 105:801–8. DOI: 10.1007/s10482-014-0128-1. PMID: 24595860. PMCID: PMC3982209.
10. Mishra AK, Kumar A, Swain DK, Yadav S, Nigam R. 2018; Insights into pH regulatory mechanisms in mediating spermatozoa functions. Vet World. 11:852–8. DOI: 10.14202/vetworld.2018.852-858. PMID: 30034181. PMCID: PMC6048093.

Go to : 




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download