Salmonella is one of the major causes of food-borne infections, including gastroenteritis with diarrhea, fever, and abdominal cramps [
1]. The United States Centers for Disease Control and Prevention (US CDC) has reported that
Salmonella causes 1.35 million illnesses, 26,500 hospitalizations, and 420 deaths in the US each year [
2]. In Korea, the prevalence of non-typhoidal
Salmonella (NTS) has been increasing, whereas that of
S. serovar Typhi has been decreasing sharply [
3,
4]. Among NTS,
S. Enteritidis and
S. Typhimurium are the most commonly isolated serotypes from clinical specimens [
5]. Recently,
Salmonella serotypes are becoming more diverse, and there are reports of outbreaks caused by unusual serotypes [
6]. Therefore, identification of the
Salmonella serotypes is required for appropriate diagnosis and treatment as well as for monitoring changes in the serotypes.
Ampicillin, chloramphenicol, and trimethoprim/sulfamethoxazole (SXT) are no longer used as primary antimicrobials because of the high resistance rates to these agents [
7]. Third-generation cephalosporin and fluoroquinolone are recommended as first-line antimicrobials; however, there have been several reports of resistance [
8,
9]. We investigated the serotype distribution and antimicrobial susceptibility of
Salmonella isolates collected in Korea between January 2016 and December 2017.
In total, 669 Salmonella isolates were collected from clinical specimens, including stool (N=456), blood (N=177), urine (N=17), pus (N=7), body fluid (N=3), sputum (N=1), and other sites (N=8), at 19 university hospitals. The geographical distribution of the specimens was as follows: Gyeongsang Province (N=168), Gyeonggi Province (N=164), Seoul (N=107), Jeolla Province (N=85), Gangwon Province (N=64), Chungcheong Province (N=43), and Jeju Province (N=38). All isolates were identified using the Vitek2 system (bioMérieux SA, Marcy L’Étoile, France) at Inje University Busan Paik Hospital. All isolates were serotyped according to the Kauffman–White scheme, using slide and tube agglutination tests with somatic (O) and flagellar (H) antisera. First, the Salmonella O group was serotyped using antiserum (Becton, Dickinson and Company, Sparks, MD, USA), and then, the H antigen was determined by confirming the coagulation reaction in Salmonella H antiserum (Becton, Dickinson and Company).
Antimicrobial susceptibility was tested using Sensititre EUVSEC susceptibility MIC plates (TREK Diagnostic Systems/Thermo Fisher Scientific, Cleveland, OH, USA). The antimicrobials tested were ampicillin, cefotaxime, ceftazidime, chloramphenicol, gentamicin, imipenem, tetracycline, ciprofloxacin, and azithromycin. SXT susceptibility was determined using disk diffusion (discs from Becton, Dickinson and Company).
Escherichia coli ATCC25922 was used for quality control. The results were interpreted according to the Clinical and Laboratory Standards Institute guidelines [
10]. Multidrug resistance (MDR) was defined as resistance to ampicillin, chloramphenicol, and SXT [
11].
Serogroup C (N=266; 39.8%) was the most common followed by B (N=245; 36.6%), D (N=135; 20.2%), G (N=6; 0.9%), A (N=4; 0.6%), E (N=4; 0.6%), K (N=3; 0.4%), M (N=3; 0.4%), I (N=1; 0.1%), X (N=1; 0.1%), and Y (N=1; 0.1%).
In total, 51 serotypes were identified. The most common serotype was
S. enterica serovar I 4,[
5],12:i:- (N=112; 16.7%), followed by
S. Enteritidis (N=108; 16.1%),
S. Bareilly (N=98; 14.6%),
S. Typhimurium (N=66; 9.9%), and
S. Infantis (N=46, 6.9%); these serotypes accounted for 64.3% of the isolates (
Table 1).
S. Agona,
S. Thompson,
S. Livingstone, and
S. Virchow accounted for 3.4% (N=23), 3.4% (N=23), 2.7% (N=18), and 2.1% (N=14), respectively.
S. Typhi (N= 20; 3.0%) and
S. Paratyphi A (N=4; 0.6%) were also isolated; however,
S. Paratyphi B and
S. Paratyphi C were not detected. Of the 669
Salmonella isolates in total, 660 were
S. enterica subspecies (ssp.) enterica, and nine were non-
S. enterica ssp.
enterica, including six
S. enterica ssp.
salamae, two
S. enterica ssp.
diarizonae, and one
S. enterica ssp.
houtenae. We identified 12 rare serotypes that were previously not reported in Korea; therefore, we believe that these are reported for the first time in clinical specimens from Korea.
Table 1
Serogroup and serotype distributions of Salmonella isolates
|
Serogroup A |
|
Serogroup C |
|
Serogroup E |
|
Serotype |
N (%) |
Serotype |
N (%) |
Serotype |
N (%) |
|
Paratyphi A |
4 (0.6%) |
|
Bareilly |
98 (14.6) |
|
Amager var 15+* |
2 (0.3) |
|
Serogroup B
|
Infantis |
46 (6.9) |
London |
1 (0.1) |
|
Serotype
|
N (%)
|
Thompson |
23 (3.4) |
Senftenberg |
1 (0.1) |
|
I4,[5],12:i:- |
112 (16.7) |
Livingstone |
18 (2.7) |
Serogroup G
|
|
Typhimurium |
66 (9.9) |
Virchow |
14 (2.1) |
Serotype
|
N (%)
|
|
Agona |
23 (3.4) |
Othmarschen |
11 (1.6) |
Telelkebir* |
3 (0.4) |
|
Saintpaul |
13 (1.9) |
Mbandaka |
10 (1.5) |
Agbeni* |
1 (0.1) |
|
Schleissheim |
7 (1.0) |
Rissen |
9 (1.3) |
NewYork* |
1 (0.1) |
|
Schwarzengrund |
6 (0.9) |
II 6,7:g,[m],s,t:(z42) |
6 (0.9) |
Poona |
1 (0.1) |
|
Stanley |
6 (0.9) |
Newport |
6 (0.9) |
Serogroup I
|
|
SanDiego |
4 (0.6) |
Braenderup |
4 (0.6) |
Serotype
|
N (%)
|
|
Derby |
3 (0.4) |
Narashino |
4 (0.6) |
Naware* |
1 (0.1) |
|
Heidelberg |
2 (0.3) |
Albany |
3 (0.4) |
Serogroup K
|
|
Coeln |
1 (0.1) |
Montevideo |
3 (0.4) |
Serotype
|
N (%)
|
|
Kaapstad* |
1 (0.1) |
Corvallis |
2 (0.3) |
Cerro* |
3 (0.4) |
|
Lagos |
1 (0.1) |
Goldcoast |
2 (0.3) |
Serogroup M
|
|
Serogroup D
|
Kentucky |
2 (0.3) |
Serotype
|
N (%)
|
|
Serotype
|
N (%)
|
Litchfield |
2 (0.3) |
Pomona |
2 (0.3) |
|
Enteritidis |
108 (16.1) |
IV 6,7:z4 z23:-* |
1 (0.1) |
Umbilo* |
1 (0.1) |
|
Typhi |
20 (3.0) |
Ferruch* |
1 (0.1) |
Serogroup X
|
|
Panama |
7 (1.0) |
Ohio |
1 (0.1) |
Serotype
|
N (%)
|
|
|
IIIb 47:r:z* |
1 (0.1) |
|
Serogroup Y
|
|
Serotype
|
N (%)
|
|
IIIb 48:k:z* |
1 (0.1) |

Table 2 lists the resistance rates of the
Salmonella isolates. The resistance rates to ampicillin, chloramphenicol, and SXT were 32.6%, 12.1%, and 8.4%, respectively. The resistance rates to cefotaxime and ceftazidime were 8.1% and 6.0%, respectively. The resistance rate to ciprofloxacin was 3.0%, and the intermediate resistance rate was high, at 24.5%. Among the nine isolates resistant to azithromycin (1.3%), three were
S. Typhi and six were NT
S.
Table 2
Antimicrobial resistance rates according to Salmonella serotype
|
Serotype |
N |
AMP |
CTX |
CAZ |
CHL |
IMI |
GEN |
TET |
CIP |
AZI |
SXT |
|
I 4,[5],12:i:- |
112 |
102 (91.1) |
16 (14.3) |
5 (4.5) |
26 (23.2) |
0 (0) |
11 (9.8) |
96 (85.7) |
7 (6.3) |
1 (0.9) |
22 (19.6) |
|
Enteritidis |
108 |
48 (44.4) |
23 (21.3) |
22 (20.4) |
27 (25.0) |
0 (0) |
18 (16.7) |
27 (25.0) |
2 (1.9) |
1 (0.9) |
8 (7.4) |
|
Bareilly |
98 |
2 (2.0) |
1 (1.0) |
1 (1.0) |
0 (0) |
0 (0) |
2 (2.0) |
2 (2.0) |
0 (0) |
0 (0) |
0 (0) |
|
Typhimurium |
66 |
35 (53.0) |
2 (3.0) |
2 (3.0) |
12 (18.2) |
0 (0) |
20 (30.3) |
31 (47.0) |
2 (3.0) |
1 (1.5) |
9 (13.6) |
|
Infantis |
46 |
1 (2.2) |
1 (2.2) |
1 (2.2) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
|
Agona |
23 |
1 (4.3) |
0 (0) |
0 (0) |
1 (4.3) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
1 (4.3) |
|
Thompson |
23 |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
|
Typhi |
20 |
0 (0) |
0 (0) |
0 (0) |
1 (5.0) |
0 (0) |
0 (0) |
3 (15.0) |
5 (25.0) |
3 (15.0) |
0 (0) |
|
Others*
|
173 |
29 (16.8) |
11 (6.4) |
9 (5.2) |
14 (8.1) |
0 (0) |
7 (4.0) |
28 (16.2) |
4 (2.3) |
3 (1.7) |
16 (9.2) |
|
Total |
669 |
218 (32.6) |
54 (8.1) |
40 (6.0) |
81 (12.1) |
0 (0) |
58 (8.7) |
187 (28.0) |
20 (3.0) |
9 (1.3) |
56 (8.4) |

The resistance rate differed depending on the serotype.
S. I 4,[
5],12:i:- had high resistance to ampicillin (91.1%), chloramphenicol (23.2%), and SXT (19.6%). The resistance rates to cefotaxime and ceftazidime were higher among
S. Enteritidis (21.3% and 20.4%) than among
S. I 4,[
5],12:i:- (14.3% and 4.5%) isolates, whereas the resistance rate to ciprofloxacin was substantially higher among
S. Typhi (25.0%) than among
S. I 4,[
5],12:i:- (6.3%) and
S. Enteritidis (1.9%) isolates. The intermediate resistance rate to ciprofloxacin was extremely high in
S. Enteritidis, at 88.0%. While
S. Bareilly had low resistance rates to all antimicrobials tested, the rate of intermediate resistance to ciprofloxacin was as high as 17.3%.
Of the 669 isolates, 5.4% (N=36) exhibited MDR. The distribution of MDR serotypes was
S. I 4,[
5],12:i:- (N=18),
S. Typhimurium (N=5), and
S. Albany (N=3) (
Fig. 1A). All three
S. Albany isolates exhibited MDR. The proportion of MDR isolates in each serotype was high in
S. Derby (1/3),
S. Panama (2/7),
S. Mbandaka (2/10),
S. Rissen (2/9), and
S. I 4,[
5],12:i:- (18/112) (
Fig. 1B). Both intermediate and resistance rates to ciprofloxacin were 69.4% and 25.1% in MDR and non-MDR isolates, respectively. The resistance rates to third-generation cephalosporins were 44.4% and 6.2% in MDR and non-MDR isolates, respectively.
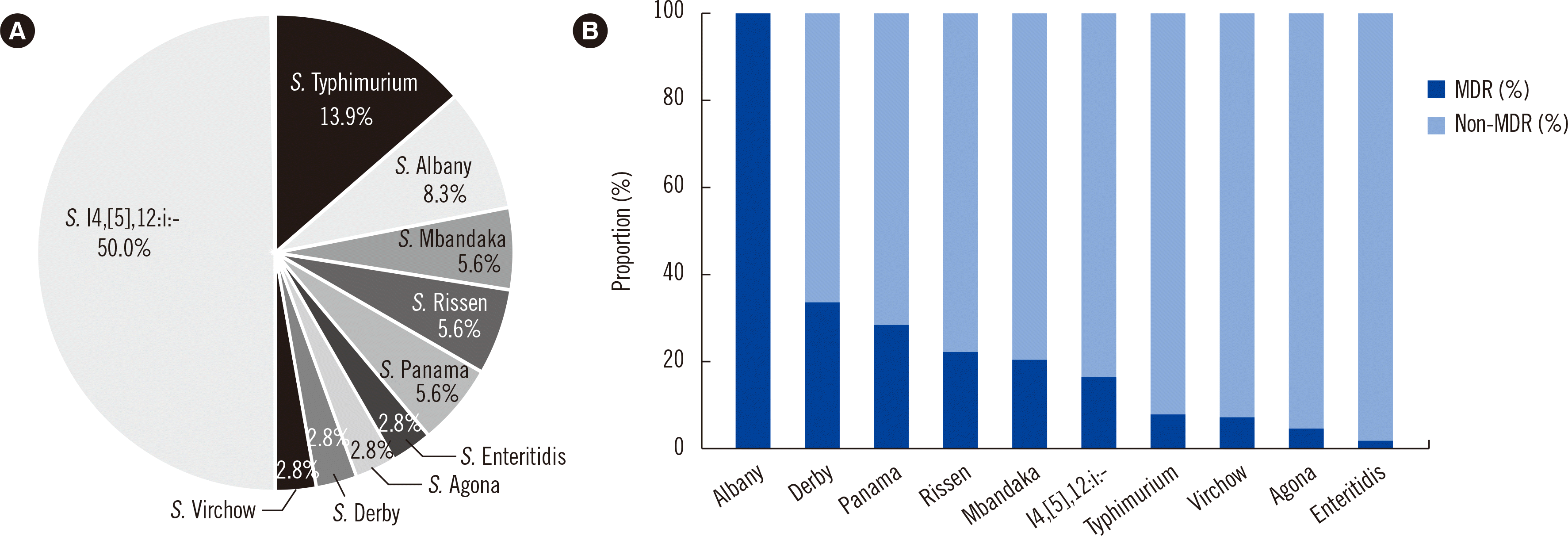 | Fig. 1
Serotype distribution of MDR Salmonella isolates collected in Korea between 2016 and 2017. (A) Distribution of MDR Salmonella isolates. (B) Proportion of MDR isolates among MDR serotypes.
Abbreviation: MDR, multidrug resistance.

|
S. Enteritidis and
S. Typhimurium were the most prevalent serotypes among NTS recovered in the past 10 yrs in Korea. Yoon,
et al. [
12] reported that the prevalence of these two serotypes was 42.0% and 21.7%, respectively, between 2004 and 2014. Accordingly, the Korea CDC reported that
S. Enteritidis (16.7%) was the most common serotype in 2014 [
13].
S. enterica I 4,[
5],12:i:- represents a monophasic variant of
S. Typhimurium, which was rarely identified until the mid-1990s; however, the prevalence of this serotype has increased recently [
14,
15]. The 2016 Annual Report by the US CDC indicated that I 4,[
5],12:i:- was the fifth most prevalent serotype and its prevalence increased by 78.3% since 2006 [
16]. In our previous study, the major serotype changed from
S. Enteritidis (16.8%) and
S. Typhimurium (12.4%) to I 4,[
5],12:i:- (23.0%) in 2015 [
17]. Similarly, in this study, the most common serotype was I 4,[
5], 12:i:- (16.9%); thus, this serotype has become the most common in Korea. In our previous studies, the prevalence of
S. Bareilly was 1.6% in 2008 and 8.0% in 2015 [
3,
17]. In this study, its prevalence increased to 14.6%; thus, we can assume that the prevalence of
S. Bareilly is steadily increasing. The prevalence of
S. Agona (3.4%) and
S. Saintpaul (1.9%) showed a slight increase compared with previous data (1.8% and 0.9%) [
17], whereas that of
S. Typhimurium (9.9%) showed a gradual decrease compared with data from 2008 (16.8%) and 2015 (12.4%) [
3,
17].
We found 12 rare serotypes that have not been previously reported as a cause of human infections in Korea. Outbreaks of food-borne diseases transmitted by livestock have been commonly reported in other countries [
18,
19]. We assume that these serotypes originated from imported foods or foreign travel, although we did not confirm the origin of the infections. Thus, continued investigation of serotype changes and the emergence of new serotypes is required.
A report of the National Antimicrobial Resistance Monitoring System revealed decreased susceptibilities to ciprofloxacin and azithromycin in 2015, although the resistance rates of NTS were <0.4% [
20]. In our previous study, we found no
Salmonella isolates resistant to either drug, although the rate of intermediate resistance to ciprofloxacin was as high as 29.2% in NTS [
17]. In this study, the resistance rate to ciprofloxacin increased to 2.2% (N=15), although the intermediate resistance rate slightly decreased among NTS isolates. Thus, there is a need to carefully monitor the increase in ciprofloxacin resistance in future.
MDR
S. Typhi has become a global concern [
11]. Thirty-six NTS isolates exhibited MDR, although there were none among
S. Typhi.
S. I 4,[
5],12:i:- was highly prevalent among all
Salmonella isolates and was the most common among MDR isolates.
In conclusion,
S. I 4,[
5],12:i:- and
S. Enteritidis are highly prevalent, and there is an increase in rare serotypes in Korea. MDR and ciprofloxacin resistance are highly prevalent. Periodic investigations of
Salmonella serotypes and antimicrobial resistance would be useful for understanding the epidemiology and for patient management.
Go to :





 PDF
PDF Citation
Citation Print
Print




 XML Download
XML Download