INTRODUCTION
MATERIALS AND METHODS
Study design
Software development
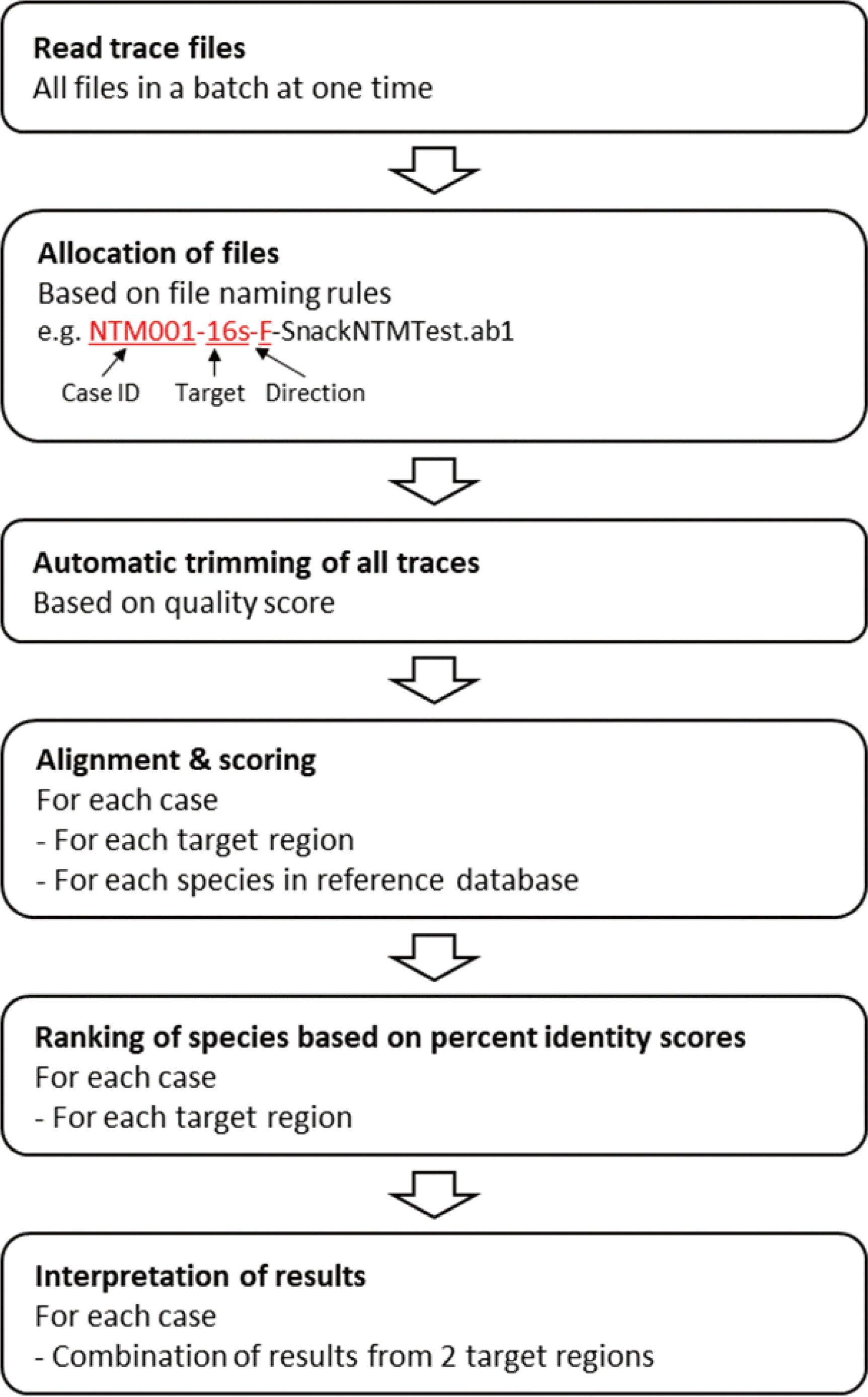 | Fig. 1Schematic representation of the workflow of SnackNTM. SnackNTM reads multiple trace files at once and allocates them according to the cases and target regions. After automatic trimming is performed for all traces, input sequences are iteratively aligned to all locally stored reference sequences. The percent identity score is then calculated for every alignment and the species are sorted according to their scores. The results for the two target regions are combined to produce the final identification result. |
Target regions and reference sequences
Identification criteria
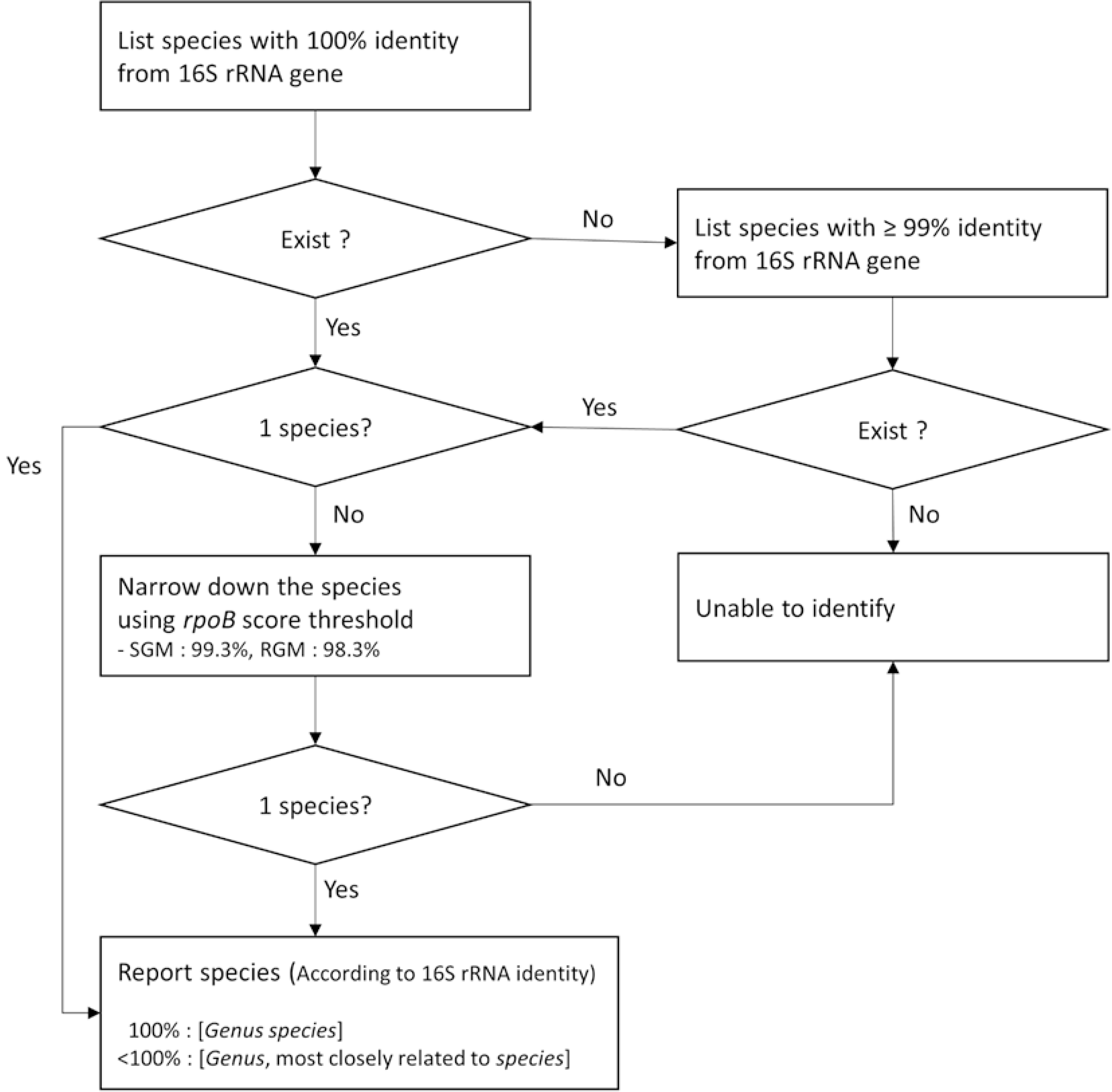 | Fig. 2Identification criteria employed in SnackNTM. Species with 100% identity based on 16S rRNA gene sequences, if available, are selected for further analysis. If not, species with ≥99% identity based on 16S rRNA gene sequences are selected. When multiple species satisfy the 16S rRNA gene sequence criteria, rpoB sequence analysis results are utilized to narrow down the species. The cutoff values used in rpoB sequence analysis were 99.3% for slowgrowing mycobacteria and 98.3% for rapidly growing mycobacteria, according to a previous study [5].
Abbreviations: SGM, slowgrowing mycobacteria; RGM, rapidly growing mycobacteria.
|
Validation using consecutive case data
Validation using unique species data
Comparison of the processing time between SnackNTM and manual identification
RESULTS
SnackNTM software
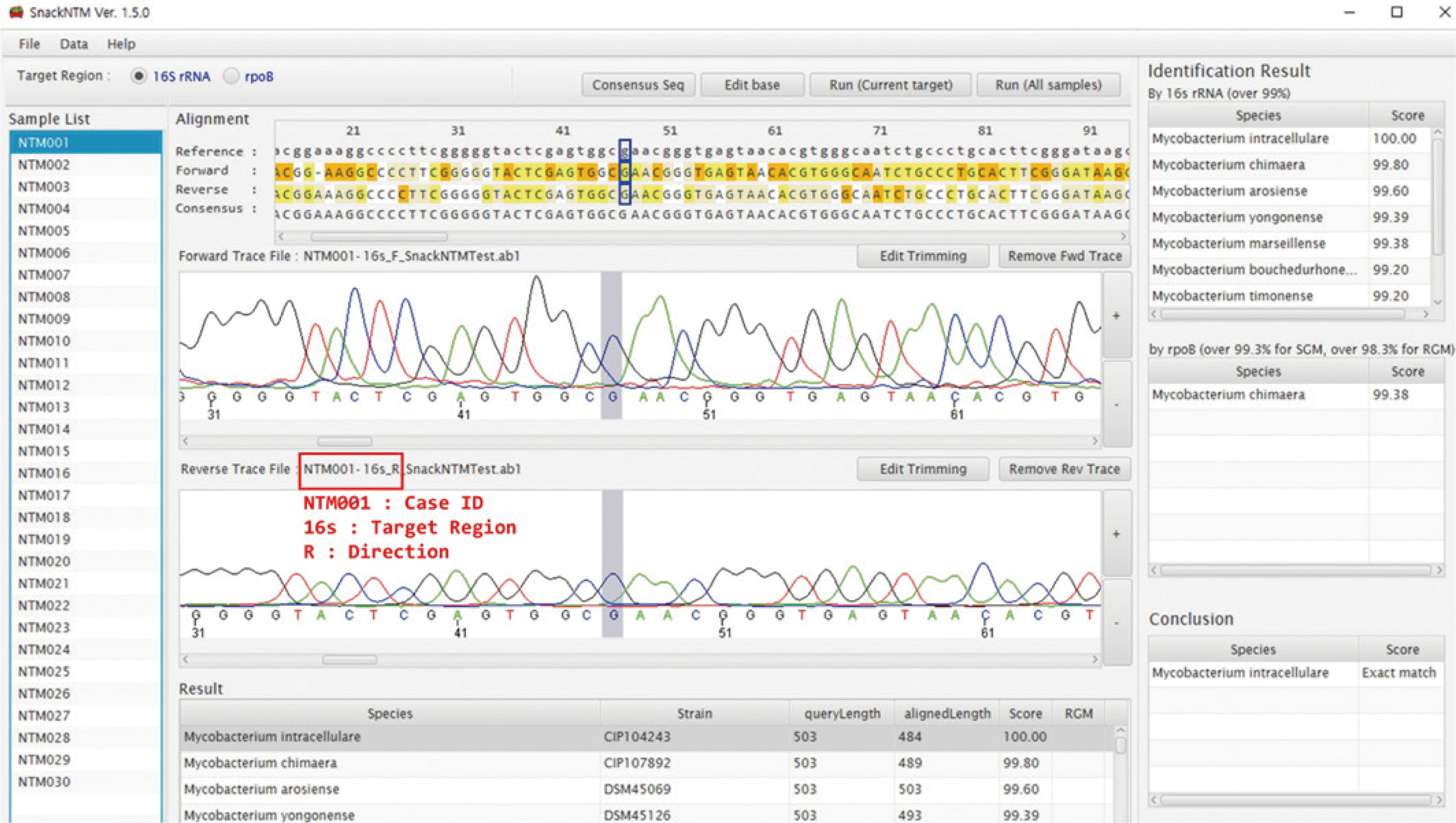 | Fig. 3Snapshot of SnackNTM after initial analysis. Trace files from all cases in a batch are loaded at once and allocated to their positions in a program according to a predefined file naming rule. The alignment results are browsed in the “Alignment” panel, and chromatograms appear below according to mouse clicks on the “Alignment” panel. Miscalled bases can be corrected by clicking the “Edit base” button. In the “Identification Result” panel, species that satisfy the criteria of each target region are shown. Final results are shown in the “Conclusion” panel. |
Validation using consecutive case data
Table 1
| Case ID | SnackNTM | Conclusion of SnackNTM | Conclusion of manual identification | Agreement* | |||
|---|---|---|---|---|---|---|---|
| 16S rRNA gene | rpoB | ||||||
| Species | % Identity | Species | % Identity | ||||
| NTM001 | M. intracellulare | 100.00 | M. chimaera | 99.38 | M. intracellulare | M. intracellulare | O |
| NTM002 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM003 | M. gastri | 99.82 | M. kansasii | 100.00 | M. kansasii | M. kansasii | O |
| M. kansasii | 99.82 | (most closely)† | (most closely) | ||||
| M. innocens | 99.47 | ||||||
| NTM004 | M. paragordonae | 99.62 | M. paragordonae | M. paragordonae | O | ||
| (most closely) | (most closely) | ||||||
| NTM005 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM006 | M. avium subsp. avium | 99.82 | M. avium subsp. avium | 99.69 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 99.81 | M. avium subsp. paratuberculosis | 99.54 | (most closely) | (most closely) | ||
| M. avium subsp. paratuberculosis | 99.62 | M. avium subsp. silvaticum | 99.38 | ||||
| M. bouchedurhonense | 99.08 | ||||||
| M. vulneris | 99.08 | ||||||
| M. colombiense | 99.03 | ||||||
| NTM007 | M. avium subsp. avium | 99.82 | M. avium subsp. avium | 99.85 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 99.81 | M. avium subsp. paratuberculosis | 99.69 | (most closely) | (most closely) | ||
| M. avium subsp. paratuberculosis | 99.63 | M. avium subsp. silvaticum | 99.54 | ||||
| M. bouchedurhonense | 99.10 | ||||||
| M. vulneris | 99.10 | ||||||
| M. colombiense | 99.05 | ||||||
| NTM008 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 100.00 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM009 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.64 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. silvaticum | 99.46 | ||||
| M. avium subsp. paratuberculosis | 99.46 | ||||||
| NTM010 | M. avium subsp. avium | 100.00 | M. avium | M. avium | O | ||
| M. avium subsp. silvaticum | 100.00 | ||||||
| NTM011 | M. avium subsp. avium | 100.00 | M. avium | M. avium | O | ||
| M. avium subsp. silvaticum | 100.00 | ||||||
| NTM012 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 100.00 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.85 | ||||
| M. avium subsp. silvaticum | 99.69 | ||||||
| NTM013 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM014 | M. avium subsp. avium | 99.82 | M. avium subsp. avium | 99.69 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 99.81 | M. avium subsp. paratuberculosis | 99.54 | (most closely) | (most closely) | ||
| M. avium subsp. paratuberculosis | 99.62 | M. avium subsp. silvaticum | 99.38 | ||||
| M. bouchedurhonense | 99.09 | ||||||
| M. colombiense | 99.03 | ||||||
| NTM015 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM016 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 100.00 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM017 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM018 | M. chelonae subsp. gwanakae | 100.00 | M. massiliense | 99.54 | M. massiliense | M. massiliense | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM019 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM020 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 100.00 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM021 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM022 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.69 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.54 | ||||
| M. avium subsp. silvaticum | 99.38 | ||||||
| NTM023 | M. avium subsp. avium | 99.82 | M. avium subsp. avium | 99.85 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 99.81 | M. avium subsp. paratuberculosis | 99.69 | (most closely) | (most closely) | ||
| M. avium subsp. paratuberculosis | 99.62 | M. avium subsp. silvaticum | 99.54 | ||||
| M. bouchedurhonense | 99.09 | ||||||
| M. colombiense | 99.03 | ||||||
| NTM024 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 100.00 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM025 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM026 | M. chelonae subsp. gwanakae | 100.00 | M. massiliense | 99.54 | M. massiliense | M. massiliense | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM027 | M. chelonae subsp. gwanakae | 100.00 | M. massiliense | 99.69 | M. massiliense | M. massiliense | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM028 | M. yongonense | 99.81 | Unidentifiable | Unidentifiable | Not Evaluable | ||
| M. marseillense | 99.81 | ||||||
| M. timonense | 99.64 | ||||||
| M. vulneris | 99.47 | ||||||
| M. chimaera | 99.44 | ||||||
| M. bouchedurhonense | 99.29 | ||||||
| M. arosiense | 99.29 | ||||||
| M. intracellulare | 99.25 | ||||||
| M. colombiense | 99.25 | ||||||
| NTM029 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.85 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.69 | ||||
| M. avium subsp. silvaticum | 99.54 | ||||||
| NTM030 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.68 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. silvaticum | 99.52 | ||||
| M. avium subsp. paratuberculosis | 99.52 | ||||||
| NTM031 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.82 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. silvaticum | 99.64 | ||||
| M. avium subsp. paratuberculosis | 99.64 | ||||||
| NTM032 | M. arcueilense | 100.00 | M. alvei | 99.38 | Unidentifiable | Unidentifiable | Not Evaluable |
| M. montmartrense | 100.00 | ||||||
| M. lutetiense | 100.00 | ||||||
| M. peregrinum | 100.00 | ||||||
| M. septicum | 100.00 | ||||||
| NTM033 | M. intracellulare | 100.00 | M. chimaera | 99.54 | M. intracellulare | M. intracellulare | O |
| NTM034 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM035 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 99.84 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM036 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.85 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.69 | ||||
| M. avium subsp. silvaticum | 99.54 | ||||||
| NTM037 | M. chelonae subsp. gwanakae | 100.00 | Unidentifiable | Unidentifiable | Not Evaluable | ||
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM038 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.69 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.54 | ||||
| M. avium subsp. silvaticum | 99.38 | ||||||
| NTM039 | M. saopaulense | 100.00 | M. massiliense | 99.69 | M. saopaulense | M. massiliense | X |
| NTM040 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.54 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.38 | ||||
| NTM041 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.69 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.53 | ||||
| M. avium subsp. silvaticum | 99.38 | ||||||
| NTM042 | M. chelonae subsp. bovis | 100.00 | M. abscessus subsp. abscessus | 100.00 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. gwanakae | 100.00 | ||||||
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM043 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.67 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.5 | ||||
| M. avium subsp. silvaticum | 99.34 | ||||||
| NTM044 | M. avium subsp. silvaticum | 100.00 | M. avium subsp. avium | 99.38 | M. avium | M. avium | O |
| M. avium subsp. avium | 100.00 | ||||||
| NTM045 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.84 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.69 | ||||
| M. avium subsp. silvaticum | 99.53 | ||||||
| NTM046 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM047 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.69 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.54 | ||||
| M. avium subsp. silvaticum | 99.38 | ||||||
| NTM048 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM049 | M. intracellulare | 100.00 | M. chimaera | 99.53 | M. intracellulare | M. intracellulare | O |
| NTM050 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.53 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.38 | ||||
| NTM051 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.52 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.36 | ||||
| NTM052 | M. intracellulare | 100.00 | M. intracellulare | 99.54 | M. intracellulare | M. intracellulare | O |
| NTM053 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM054 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM055 | M. mageritense | 100.00 | M. mageritense | M. mageritense | O | ||
| NTM056 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM057 | M. avium subsp. avium | 100.00 | M. avium | M. avium | O | ||
| M. avium subsp. silvaticum | 100.00 | ||||||
| NTM058 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.69 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.53 | ||||
| M. avium subsp. silvaticum | 99.37 | ||||||
| NTM059 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM060 | M. intracellulare | 100.00 | M. intracellulare | 99.85 | M. intracellulare | M. intracellulare | O |
| M. chimaera | 99.54 | ||||||
| NTM061 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.68 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.53 | ||||
| M. avium subsp. silvaticum | 99.37 | ||||||
| NTM062 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM063 | M. chelonae subsp. bovis | 100.00 | M. abscessus subsp. abscessus | 100.00 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. gwanakae | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM064 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM065 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.69 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.54 | ||||
| M. avium subsp. silvaticum | 99.38 | ||||||
| NTM066 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM067 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.84 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.69 | ||||
| M. avium subsp. silvaticum | 99.53 | ||||||
| NTM068 | M. chelonae subsp. gwanakae | 100.00 | M. massiliense | 99.53 | M. massiliense | M. massiliense | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM069 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.69 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.54 | ||||
| M. avium subsp. silvaticum | 99.39 | ||||||
| NTM070 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM071 | M. intracellulare | 100.00 | M. chimaera | 99.37 | M. intracellulare | M. intracellulare | O |
| NTM072 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM073 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 99.84 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM074 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 100.00 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM075 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 100.00 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM076 | M. gastri | 100.00 | M. kansasii | 99.84 | M. kansasii | M. kansasii | O |
| M. kansasii | 100.00 | ||||||
| NTM077 | M. avium subsp. avium | 100.00 | M. avium | M. avium | O | ||
| M. avium subsp. silvaticum | 100.00 | ||||||
| NTM078 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM079 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM080 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM081 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM082 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM083 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM084 | M. avium subsp. avium | 100.00 | M. avium | M. avium | O | ||
| M. avium subsp. silvaticum | 100.00 | ||||||
| NTM085 | M. chelonae subsp. gwanakae | 100.00 | Unidentifiable | Unidentifiable | Not Evaluable | ||
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM086 | M. chimaera | 99.81 | M. chimaera | 100.00 | Unidentifiable | Unidentifiable | Not Evaluable |
| M. intracellulare | 99.62 | M. intracellulare | 99.67 | ||||
| M. yongonense | 99.44 | ||||||
| M. marseillense | 99.44 | ||||||
| M. timonense | 99.29 | ||||||
| M. arosiense | 99.29 | ||||||
| NTM087 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM088 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 100.00 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM089 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM090 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.54 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.39 | ||||
| NTM091 | M. intracellulare | 100.00 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O |
| NTM092 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.85 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.69 | ||||
| M. avium subsp. silvaticum | 99.54 | ||||||
| NTM093 | M. chelonae subsp. gwanakae | 100.00 | Unidentifiable | Unidentifiable | Not Evaluable | ||
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM094 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 99.68 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM095 | M. gastri | 100.00 | Unidentifiable | Unidentifiable | Not Evaluable | ||
| M. kansasii | 100.00 | ||||||
| NTM096 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 99.68 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM097 | M. intracellulare | 100.00 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O |
| M. chimaera | 99.69 | ||||||
| NTM098 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM099 | M. avium subsp. avium | 100.00 | M. avium | M. avium | O | ||
| M. avium subsp. silvaticum | 100.00 | ||||||
| NTM100 | M. intracellulare | 100.00 | M. chimaera | 99.38 | M. intracellulare | M. intracellulare | O |
| NTM101 | M. chelonae subsp. gwanakae | 100.00 | M. massiliense | 99.68 | M. massiliense | M. massiliense | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM102 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.84 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. silvaticum | 99.67 | ||||
| M. avium subsp. paratuberculosis | 99.67 | ||||||
| NTM103 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.68 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.52 | ||||
| M. avium subsp. silvaticum | 99.37 | ||||||
| NTM104 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.84 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.68 | ||||
| M. avium subsp. silvaticum | 99.52 | ||||||
| NTM105 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM106 | M. chelonae subsp. gwanakae | 100.00 | M. massiliense | 99.67 | M. massiliense | M. massiliense | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM107 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.84 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.68 | ||||
| M. avium subsp. silvaticum | 99.52 | ||||||
| NTM108 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM109 | M. intracellulare | 100.00 | M. chimaera | 99.52 | M. intracellulare | M. intracellulare | O |
| NTM110 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 99.69 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM111 | M. intracellulare | 100.00 | M. chimaera | 99.52 | M. intracellulare | M. intracellulare | O |
| NTM112 | M. paragordonae | 99.81 | M. paragordonae | M. paragordonae | O | ||
| (most closely) | (most closely) | ||||||
| NTM113 | M. avium subsp. avium | 100.00 | M. avium | M. avium | O | ||
| M. avium subsp. silvaticum | 100.00 | ||||||
| NTM114 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.69 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.54 | ||||
| M. avium subsp. silvaticum | 99.39 | ||||||
| NTM115 | M. chelonae subsp. gwanakae | 100.00 | M. massiliense | 99.62 | M. massiliense | M. massiliense | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM116 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.69 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.54 | ||||
| M. avium subsp. silvaticum | 99.38 | ||||||
| NTM117 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.8 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. silvaticum | 99.6 | ||||
| M. avium subsp. paratuberculosis | 99.6 | ||||||
| NTM118 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.38 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | ||||||
| NTM119 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 100.00 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM120 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM121 | M. avium subsp. avium | 100.00 | M. avium | M. avium | O | ||
| M. avium subsp. silvaticum | 100.00 | ||||||
| NTM122 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM123 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.83 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.66 | ||||
| M. avium subsp. silvaticum | 99.48 | ||||||
| NTM124 | M. bouchedurhonense | 99.45 | M. avium subsp. avium | 99.84 | M. avium | M. avium | O |
| M. avium subsp. avium | 99.45 | M. avium subsp. silvaticum | 99.68 | (most closely) | (most closely) | ||
| M. avium subsp. silvaticum | 99.42 | M. avium subsp. paratuberculosis | 99.68 | ||||
| M. colombiense | 99.42 | ||||||
| M. avium subsp. paratuberculosis | 99.25 | ||||||
| M. vulneris | 99.08 | ||||||
| M. arosiense | 99.08 | ||||||
| M. intracellulare | 99.03 | ||||||
| NTM125 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 100.00 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. silvaticum | 99.84 | ||||
| M. avium subsp. paratuberculosis | 99.84 | ||||||
| NTM126 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.66 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. silvaticum | 99.48 | ||||
| M. avium subsp. paratuberculosis | 99.48 | ||||||
| NTM127 | M. intracellulare | 100.00 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O |
| M. chimaera | 99.70 | ||||||
| NTM128 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.85 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.69 | ||||
| M. avium subsp. silvaticum | 99.54 | ||||||
| NTM129 | M. avium subsp. avium | 99.81 | M. avium subsp. avium | 99.69 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 99.80 | M. avium subsp. paratuberculosis | 99.54 | (most closely) | (most closely) | ||
| M. avium subsp. paratuberculosis | 99.61 | M. avium subsp. silvaticum | 99.38 | ||||
| M. bouchedurhonense | 99.05 | ||||||
| NTM130 | M. intracellulare | 100.00 | M. chimaera | 99.53 | M. intracellulare | M. intracellulare | O |
| NTM131 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM132 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM133 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 100.00 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM134 | M. chelonae subsp. gwanakae | 100.00 | M. massiliense | 99.68 | M. massiliense | M. massiliense | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM135 | M. intracellulare | 100.00 | M. chimaera | 99.54 | M. intracellulare | M. intracellulare | O |
| NTM136 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.85 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.69 | ||||
| M. avium subsp. silvaticum | 99.54 | ||||||
| NTM137 | M. saopaulense | 100.00 | M. abscessus subsp. abscessus | 100.00 | M. saopaulense | M. abscessus subsp. abscessus | X |
| NTM138 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.85 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.69 | ||||
| M. avium subsp. silvaticum | 99.54 | ||||||
| NTM139 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 100.00 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM140 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 99.84 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM141 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.65 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.48 | ||||
| M. avium subsp. silvaticum | 99.31 | ||||||
| NTM142 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM143 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 99.69 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM144 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.85 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.69 | ||||
| M. avium subsp. silvaticum | 99.54 | ||||||
| NTM145 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.83 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.66 | ||||
| M. avium subsp. silvaticum | 99.50 | ||||||
| NTM146 | M. intracellulare | 100.00 | M. intracellulare | 99.37 | M. intracellulare | M. intracellulare | O |
| M. chimaera | 99.37 | ||||||
| NTM147 | M. chelonae subsp. gwanakae | 100.00 | M. chelonae subsp. chelonae | 99.69 | M. chelonae | M. chelonae | O |
| M. chelonae subsp. chelonae | 100.00 | M. chelonae subsp. gwanakae | 99.38 | ||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM148 | M. intracellulare | 100.00 | M. chimaera | 99.39 | M. intracellulare | M. intracellulare | O |
| NTM149 | M. intracellulare | 100.00 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O |
| M. chimaera | 99.69 | ||||||
| NTM150 | M. avium subsp. silvaticum | 100.00 | M. avium | M. avium | O | ||
| M. avium subsp. avium | 100.00 | ||||||
| NTM151 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.85 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.69 | ||||
| M. avium subsp. silvaticum | 99.54 | ||||||
| NTM152 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM153 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 100.00 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.84 | ||||
| M. avium subsp. silvaticum | 99.69 | ||||||
| NTM154 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 99.68 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM155 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.84 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.68 | ||||
| M. avium subsp. silvaticum | 99.52 | ||||||
| NTM156 | M. intracellulare | 100.00 | M. chimaera | 99.54 | M. intracellulare | M. intracellulare | O |
| NTM157 | M. avium subsp. avium | 100.00 | M. avium | M. avium | O | ||
| M. avium subsp. silvaticum | 100.00 | ||||||
| NTM158 | M. chelonae subsp. gwanakae | 100.00 | M. massiliense | 99.54 | M. massiliense | M. massiliense | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM159 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.68 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.52 | ||||
| M. avium subsp. silvaticum | 99.37 | ||||||
| NTM160 | M. fortuitum subsp. fortuitum | 100.00 | M. fortuitum | M. fortuitum | O | ||
| M. fortuitum subsp. acetamidolyticum | 100.00 | ||||||
| NTM161 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 100.00 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM162 | M. chimaera | 100.00 | M. chimaera | 99.85 | M. chimaera | M. chimaera | O |
| M. intracellulare | 99.54 | ||||||
| NTM163 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM164 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM165 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM166 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.85 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.69 | ||||
| M. avium subsp. silvaticum | 99.54 | ||||||
| NTM167 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM168 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.69 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.54 | ||||
| M. avium subsp. silvaticum | 99.38 | ||||||
| NTM169 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 100.00 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM170 | M. chelonae subsp. gwanakae | 100.00 | M. massiliense | 99.69 | M. massiliense | M. massiliense | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM171 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 100.00 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM172 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.85 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.69 | ||||
| M. avium subsp. silvaticum | 99.54 | ||||||
| NTM173 | M. intracellulare | 100.00 | M. intracellulare | 99.85 | M. intracellulare | M. intracellulare | O |
| M. chimaera | 99.85 | ||||||
| NTM174 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 100.00 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.85 | ||||
| M. avium subsp. silvaticum | 99.69 | ||||||
| NTM175 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 100.00 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM176 | M. chelonae subsp. gwanakae | 100.00 | M. massiliense | 99.54 | M. massiliense | M. massiliense | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM177 | M. chelonae subsp. gwanakae | 100.00 | M. massiliense | 99.37 | M. massiliense | M. massiliense | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM178 | M. arcueilense | 100.00 | M. alvei | 99.38 | Unidentifiable | Unidentifiable | Not Evaluable |
| M. montmartrense | 100.00 | ||||||
| M. lutetiense | 100.00 | ||||||
| M. peregrinum | 100.00 | ||||||
| M. septicum | 100.00 | ||||||
| NTM179 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM180 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.84 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.68 | ||||
| M. avium subsp. silvaticum | 99.52 | ||||||
| NTM181 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 100.00 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.85 | ||||
| M. avium subsp. silvaticum | 99.69 | ||||||
| NTM182 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.69 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.54 | ||||
| M. avium subsp. silvaticum | 99.39 | ||||||
| NTM183 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.69 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.54 | ||||
| M. avium subsp. silvaticum | 99.38 | ||||||
| NTM184 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.85 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.69 | ||||
| M. avium subsp. silvaticum | 99.54 | ||||||
| NTM185 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.69 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.54 | ||||
| M. avium subsp. silvaticum | 99.39 | ||||||
| NTM186 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM187 | M. chelonae subsp. gwanakae | 100.00 | M. massiliense | 99.69 | M. massiliense | M. massiliense | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM188 | M. intracellulare | 100.00 | M. intracellulare | 99.85 | M. intracellulare | M. intracellulare | O |
| M. chimaera | 99.85 | ||||||
| NTM189 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.69 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.54 | ||||
| M. avium subsp. silvaticum | 99.39 | ||||||
| NTM190 | M. intracellulare | 100.00 | M. chimaera | 99.51 | M. intracellulare | M. intracellulare | O |
| NTM191 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM192 | M. chelonae subsp. gwanakae | 100.00 | Unidentifiable | Unidentifiable | Not Evaluable | ||
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM193 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM194 | M. chelonae subsp. gwanakae | 100.00 | M. massiliense | 99.53 | M. massiliense | M. massiliense | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM195 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM196 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.67 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. silvaticum | 99.51 | ||||
| M. avium subsp. paratuberculosis | 99.51 | ||||||
| NTM197 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.69 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.54 | ||||
| M. avium subsp. silvaticum | 99.39 | ||||||
| NTM198 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 100.00 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM199 | M. porcinum | 100.00 | M. porcinum | 100.00 | M. porcinum | M. porcinum | O |
| M. neworleansense | 100.00 | ||||||
| NTM200 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM201 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM202 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM203 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.85 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.69 | ||||
| M. avium subsp. silvaticum | 99.54 | ||||||
| NTM204 | M. chelonae subsp. gwanakae | 100.00 | M. massiliense | 99.56 | M. massiliense | M. massiliense | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM205 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.69 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.54 | ||||
| M. avium subsp. silvaticum | 99.38 | ||||||
| NTM206 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.84 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. silvaticum | 99.67 | ||||
| M. avium subsp. paratuberculosis | 99.67 | ||||||
| NTM207 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.83 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. silvaticum | 99.67 | ||||
| M. avium subsp. paratuberculosis | 99.67 | ||||||
| NTM208 | M. chelonae subsp. gwanakae | 100.00 | M. massiliense | 99.83 | M. massiliense | M. massiliense | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM209 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM210 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 99.67 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM211 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM212 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.69 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.54 | ||||
| M. avium subsp. silvaticum | 99.38 | ||||||
| NTM213 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O | ||
| NTM214 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 100.00 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. paratuberculosis | 99.84 | ||||
| M. avium subsp. silvaticum | 99.69 | ||||||
| NTM215 | M. chelonae subsp. gwanakae | 100.00 | M. abscessus subsp. abscessus | 99.84 | M. abscessus subsp. abscessus | M. abscessus subsp. abscessus | O |
| M. chelonae subsp. chelonae | 100.00 | ||||||
| M. abscessus subsp. abscessus | 100.00 | ||||||
| M. massiliense | 100.00 | ||||||
| M. abscessus subsp. bolletii | 100.00 | ||||||
| M. chelonae subsp. bovis | 100.00 | ||||||
| M. stephanolepidis | 100.00 | ||||||
| M. franklinii | 100.00 | ||||||
| M. saopaulense | 100.00 | ||||||
| NTM216 | M. avium subsp. avium | 100.00 | M. avium subsp. avium | 99.84 | M. avium | M. avium | O |
| M. avium subsp. silvaticum | 100.00 | M. avium subsp. silvaticum | 99.67 | ||||
| M. avium subsp. paratuberculosis | 99.67 | ||||||
| NTM217 | M. intracellulare | 100.00 | M. intracellulare | 100.00 | M. intracellulare | M. intracellulare | O |
| M. chimaera | 99.69 | ||||||
*O: agreement between SnackNTM and manual identification, X: discrepancy between SnackNTM and manual identification. †According to the identification criteria described in Fig. 2, an identification result that did not show 100% identity for the 16S rRNA sequence was designated as “most closely.”




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download