Spinocerebellar ataxia type 1 (SCA1) is an autosomal dominant disease characterized by progressive cerebellar ataxia, dysarthria, and, eventually, deterioration of bulbar functions [
1]. The prevalence of SCA1 is approximately 1–2/100,000, and SCA1 accounts for approximately 6% of patients with autosomal dominant cerebellar ataxia worldwide, although the proportions vary with population [2]. SCA1 is caused by an abnormal CAG repeat expansion in the ataxin 1 gene (
ATXN1). Normal alleles have a repeat number ranging from 6–44, with 1–3 CAT interruption(s); approximately 78% of normal
ATXN1 alleles have a configuration of (CAG)nCATCAGCAT(CAG)n, 11% have (CAG)nCAT(CAG)n, and 4% have (CAG)nCATCAGCATCAGCAT(CAG)n [3]. Since there is an overlapping CAG repeat range (36–44 repeats) in healthy individuals and SCA1 patients, the presence or absence of CAT interruption(s) is important for interpreting the allele status within that range: CAT interruption indicates a normal allele, whereas in the case of no interruption, the allele within the 36–38 repeat range is interpreted as mutable normal, and the allele within the 39–44 repeat range is interpreted as full mutation [4]. The CAT interruption status might also be a relevant indicator for patients with a full mutation allele with >44 repeats, as the length of the longest uninterrupted CAG repeat stretch is correlated with age on SCA1 onset [5-7].
As PCR with flanking primers followed by fluorescent fragment length analysis cannot determine the presence or absence of an interrupted sequence, PCR with restriction enzyme digestion has been used for CAT interruption detection in ATXN1 alleles [3]. Repeat-primed (RP)-PCR and tethering PCR can also recognize interruption(s) based on a characteristic electropherogram pattern [8-10]. Despite some reports on Korean SCA1 patients [11-13], little is known about the CAT interruption status of expanded alleles and the proportion of 39–44 repeat alleles among expanded alleles. As SCA1 is rare and the probability of encountering expanded alleles is very low, it is difficult to implement a routine diagnostic testing for CAT interruption(s). We compared the traditional restriction enzyme method and fluorescence-based tethering PCR approach to detect CAT interruptions in archived SCA1 patient samples to develop a new strategy combining these two methods as a diagnostic test in Korean patients. The Institutional Review Board of Samsung Medical Center, Seoul, Korea approved this study and waived the need for informed consent (IRB No. 2020-12-003).
In total, 1,156 patients were tested for SCA1 as part of the differential diagnosis of autosomal dominant spinocerebellar ataxia at the Samsung Medical Center between January 2010 and December 2019 using fluorescent fragment analysis with flanking primers [5´ carboxyfluorescein (FAM)-CCAGACGCCGGGACACA-3´ and 5´-CCGGAGCCCTGCTGAGGT-3´] [4]. Among the 2,312 alleles, 12 alleles (0.5%) harbored 39–44 repeats, and five alleles (0.2%) harbored >44 repeats (
Fig. 1).
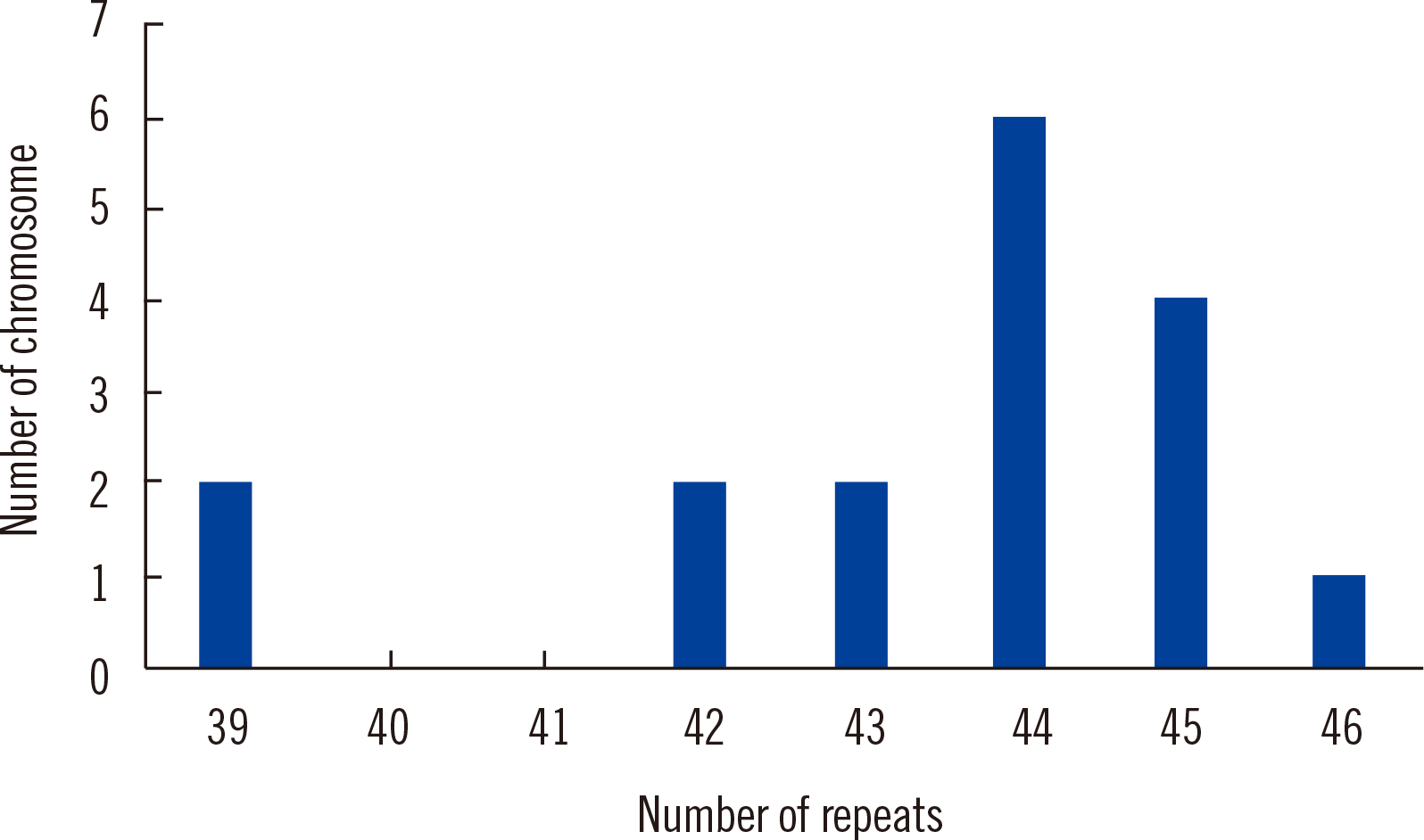 | Fig. 1Distribution of the number of repeats of ATXN1 alleles with ≥39 repeats. 
|
We performed SfaNI restriction enzyme digestion analysis of DNA samples from one healthy subject, three proficiency test materials from Korean Institute of Genetic Testing Evaluation, and six patients with ≥39 repeats archived as positive controls and stored at –20°C within two years from the start of the study. The number of repeats in the expanded allele varied across among patient samples: 42 (one sample), 43 (two samples), 44 (one sample), 45 (one sample), and 46 (one sample). PCR with primers 5´-AACTGGAAATGTGGACGTAC-3´ and 5´-CAACATGGGCAGTCTGAG-3´ followed by SfaNI enzyme digestion was performed as previously described [5]. As the SfaNI enzyme recognizes and cleaves the CAT sequence, the normal allele is expected to be cleaved, whereas the full-penetrance allele is not. Expanded alleles in the patient and proficiency test samples harbored no CAT interruption(s), whereas normal alleles did (
Fig. 2), which indicated that the expanded alleles were full-penetrance alleles.
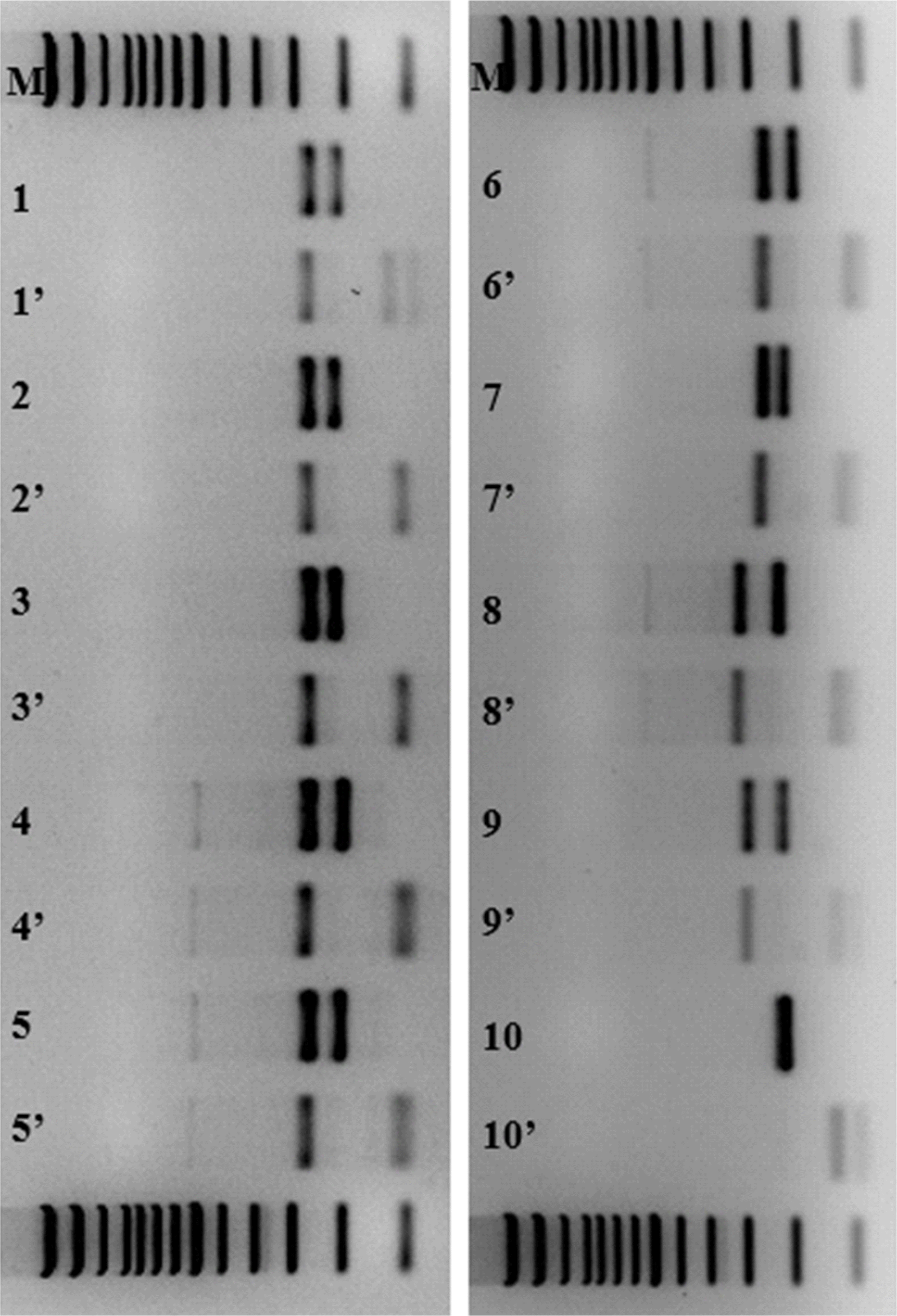 | Fig. 2Electrophoresis of PCR products before and after digestion with SfaNI enzyme. Lane M, size marker; lanes 1–6, patient samples with 43, 45, 44, 46, 43, and 42 repeats in the expanded allele, respectively; lanes 7–9, proficiency test materials with 43, 60, and 52 repeats in the expanded allele, respectively; lane 10, negative control. Lanes labeled N and N’ indicate PCR products before and after enzyme digestion, respectively. 
|
Tethering PCR was performed on 10 DNA samples that were tested for SfaNI restriction enzyme digestion analysis, using a locus-specific fluorescently labeled forward primer (5´-FAM-TTTGCTGGAGGCCTATTCCACTCT-3´) and reverse primer (5´-GAGCCCTGCTGAGGTGCTGCTGCTGCTGCTG-3´), as previously described [10]. The reverse primer contained 16 bases of a locus-specific sequence, followed by five CTG units complementary to the (CAG)n repeats. Amplicons were loaded on an ABI Prism 3730XL system (Thermo Fisher Scientific, Foster City, CA, USA) with the GS-500 size standard. Data were processed using Genemarker software version 1.95 (SoftGenetics, State College, PA, USA). Besides the main peaks caused by annealing of the reverse primer to perfectly matched bases, a stutter pattern with a 3-bp interval was obtained due to random annealing of the reverse primer within CAG repeats via the five CTG units. The first stutter peak indicated a five-CAG repeat, and the number of repeats were estimated by counting the following peaks. The presence of CAT interruption(s) was ere recognized by the loss of or reduction in signal intensity, as the reverse primer cannot bind to interrupted CAT sequences. All nine expanded alleles among the 20 alleles from the 10 DNA samples showed the expected pattern.
Fig. 3 shows an electropherogram of CAT interruption patterns for representative samples. For normal homozygous samples, there was an area without a fluorescence signal due to an interrupted sequence, which the reverse primer containing the CAG repeat cannot bind to. In SCA1 patient samples with an uninterrupted expanded allele and a normal interrupted allele, there is an area where the peak height abruptly decreases.
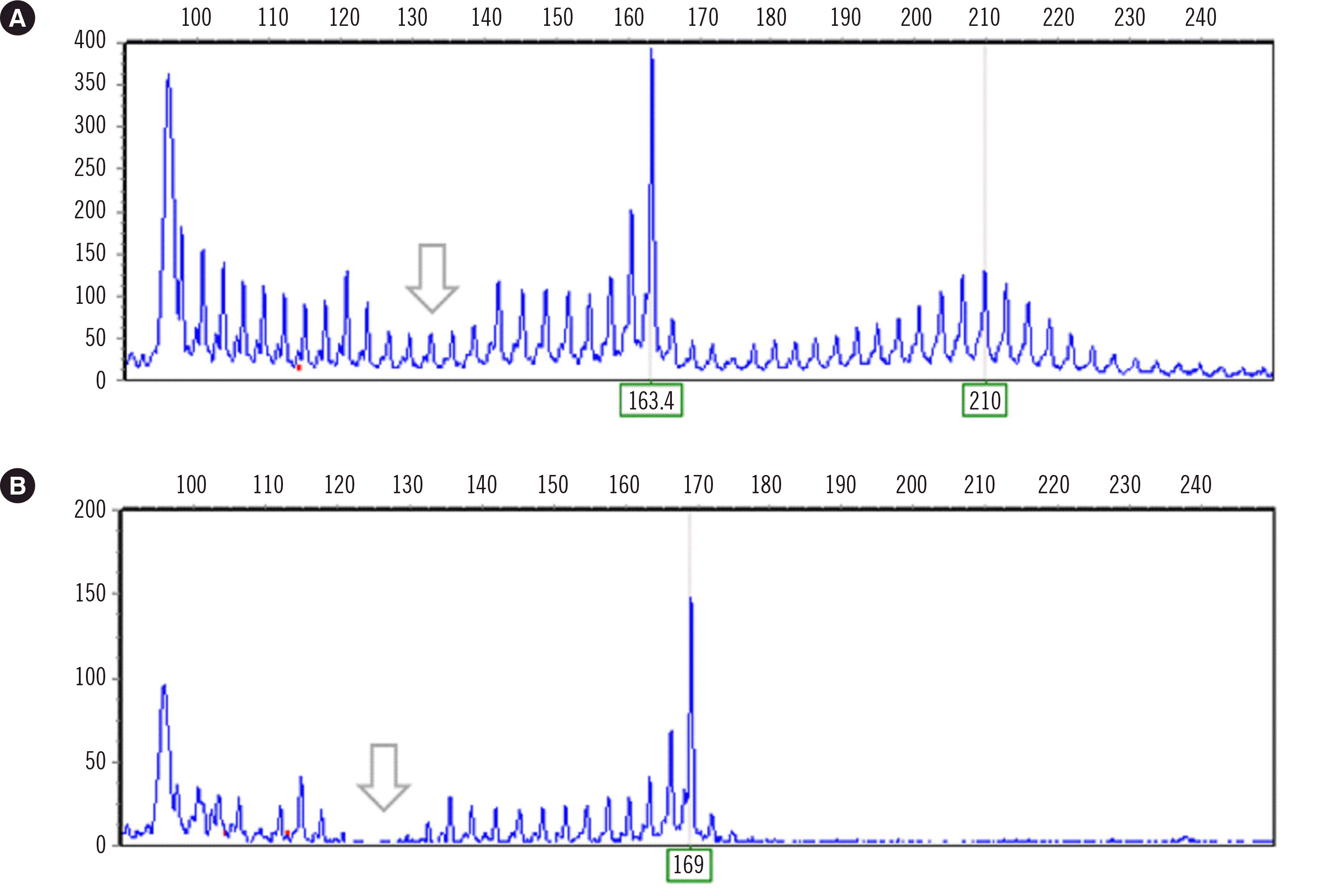 | Fig. 3Electropherograms of fluorescent fragment length analysis. (A) Electropherogram pattern of a sample with a normal interrupted (26 repeats) and an expanded uninterrupted allele (42 repeats). (B) Electropherogram pattern of a sample with a normal homozygous allele (28 repeats) with CAT interruption(s) in both alleles. The arrow indicates a reduction in signal due to the presence of interruption(s) in the normal allele. 
|
The proportion of alleles with ≥39 repeats was similar (0.862%) to that reported in a previous UK study that analyzed 6,378 SCA1 alleles [14]; however, the proportion of the 39–44 repeat alleles among expanded alleles was higher in the present study (72% vs. 36%). SfaNI restriction enzyme digestion confirmed that all tested patient samples with expanded alleles had uninterrupted CAG repeats. Among the four patients with 39–44 repeat alleles, which were confirmed to have uninterrupted CAG stretches, clinical information was available for three patients, and their age at disease onset ranged from 44 to 59 years. Onset in these patients seemed to be rather late, considering that the reported mean age of SCA1 onset is in the fourth decade of life [15, 16]. However, our results are comparable to a previous report on intermediate alleles, where the mean age of SCA1 onset among patients with uninterrupted 39–44 repeat alleles was 62 years (range: 49–85 years) [17].
The present study confirms the lack of CAT interruption(s) in ATXN1 in Korean SCA1 patients. However, follow-up studies are needed since these results are based on a small sample size. Tethering PCR may be a convenient method for clinical laboratories as it allows determining the size of alleles and identifying the presence or absence of CAT interruption(s) based on a loss or reduction in fluorescence intensity. However, it does not reveal whether or not the interruption(s) is(are) present in the expanded allele. If a stutter pattern indicates the presence of interruption(s) in one allele, further restriction enzyme tests are needed to determine whether the interruption(s) is(are) present in the normal or expanded allele.
Go to :







 PDF
PDF Citation
Citation Print
Print




 XML Download
XML Download