INTRODUCTION
Extraintestinal pathogenic
Escherichia coli (ExPEC) causes various infections, including urinary tract infection (UTI), sepsis, and neonatal meningitis [
1,
2]. The mechanisms underlying ExPEC transmission and the selection of resistant clones are poorly understood.
Among multidrug-resistant ExPEC strains, the most frequent sequence type (ST) is ST131, which is globally disseminated and resistant to multiple antibiotics [
3,
4]. Recently, ST1193, which is resistant to fluoroquinolones, has spread rapidly [
4]. ExPEC strains have virulence factors (VFs), including adhesion molecules, iron acquisition systems, invasion proteins, and toxins, which facilitate infection by allowing bacterial cells to migrate into and multiply within the host. Several ExPEC VF genes, including
pap,
vat,
kpsMII,
ibeA, and
clbB/N, are significantly associated with the dissemination of ExPEC isolates [
5,
6]. The yersiniabactin siderophore receptor encoded by
fyuA is involved in the efficient uptake of iron from the bloodstream and invasion of the bloodstream from the urinary tract [
7]. Pneumonia-specific
E. coli isolates carry higher proportions of the VF genes
sfa/foc,
papGIII,
hlyC,
cnf1, and
iroN than bacteremia isolates [
8]. The gene
cnf1 encodes cytotoxic necrotizing factor 1, a toxin associated with sepsis severity, whereas the presence of
fyuA is associated with mortality. Further, the presence of P fimbriae is associated with improved bacterial survival [
9,
10]. The presence of hek/hra may predict clinical outcomes, as one study showed an association between the presence of these genes and mortality in newborns [
11].
ExPEC strains involved in UTIs are believed to express a diverse repertoire of VFs to colonize and infect the urinary tract in an ascending manner [
1]. However, the role of VFs in the pathogenesis and clinical outcomes of
E. coli bacteremia remains to be investigated. We hypothesized that some VFs of ExPEC are involved in bacterial movement into the bloodstream in UTI patients, resulting in urosepsis. Therefore, we compared the repertoire of STs and VF genes between isolates from urine and blood from UTI patients using whole-genome sequencing (WGS) analysis.
Go to :

DISCUSSION
We hypothesized that we could define specific VFs of E. coli linked to bacterial movement from the urinary tract to the circulation. However, virulence properties as well as molecular epidemiological traits and antimicrobial resistance patterns showed no significant differences between blood and urine isolates from our cohort of UTI patients, suggesting that specific VFs of E. coli do not contribute to bacterial dissemination from the urinary tract to the circulation. However, specific isolates, such as ST131, carried more VF genes and exhibited higher antimicrobial resistance than the other isolates evaluated.
We classified isolates according to ST-
fimH-OH serotype clonal groups using WGS. The ST131-
fimH30-O25:H4 type was the most predominant and was highly resistant to several antibiotics, including β-lactams and fluoroquinolones. Among the 14 ST131-
fimH30-O25:H4 isolates, 12 produced CTX-M type EBSLs and carried variations in
gyrA,
parC, and
parE. The next three most prevalent ST-
fimH isolates were ST95-
fimH27, ST69-
fimH27, and ST1193-
fimH64. ST131 is the predominant
E. coli lineage among multidrug-resistant ExPEC isolates worldwide [
14]. Most ST131 isolates are fluoroquinolone-resistant, many are co-resistant to aminoglycosides and cotrimoxazole, and a minority produce ESBLs, such as CTX-M enzymes [
5,
14]. In our study, 78.9% of the ST131 isolates produced CTX-M ESBLs and were resistant to fluoroquinolones, and 52.6% of these were co-resistant to aminoglycosides and cotrimoxazole. After the rapid expansion of ST131
E. coli strains, fluoroquinolone-resistant ST1193 isolates emerged [
4]. ST1193 strains, derived from ST14, are commonly resistant to fluoroquinolones.
fimH64 is a highly specific marker for ST1193, a clonal group that may be entirely fluoroquinolone-resistant [
4]. Consistent herewith, all ST1193-
fimH64 isolates in our study were resistant to fluoroquinolones.
Adhesins, including fimbriae and afimbrial adhesins, play a significant role in host cell colonization [
7,
15]. We detected type I fimbriae,
E. coli common pilus, hemorrhagic
E. coli pilus, and EaeH in all isolates. P fimbriae were detected in 80% of the isolates. ExPEC can cause sepsis and infections under very low iron availability, as this pathotype has developed many strategies for obtaining iron [
16]. The sitABCD system is a membrane pump system, and the ChuA transporter enables Fe uptake directly from extracellular heme. In addition, ExPEC have siderophores, such as salmochelin, yersiniabactin, and aerobactin, which are small molecules with high affinity for Fe ions that indirectly uptake Fe [
17]. Yersiniabactin contributes to the pathogenicity of UPEC during urinary tract colonization [
18,
19]. In our study, 98% isolates had yersiniabactin, the sitABCD system, and the chuA transporter. Aerobactin receptors are substantially more efficient at capturing Fe than enterobactin receptors [
7]. In this study, 74% and 89% of the ST131 and ST1193 isolates, respectively, had the gene encoding aerobactin. An ExPEC salmochelin marker gene,
iroN, was detected in 5% of the ST131 isolates, but in none of the ST1193 isolates.
Some of the most frequently detected toxin genes in ExPEC are
hlyA,
hlyD,
hlyF,
cdtB,
tsh,
sat,
pic,
vat, and
astA [
7].
hlyF,
cdtB, and
tsh have been detected in NMEC strains [
20]. While all isolates carried hlyE in this study, alpha-hemolysins encoded by
hlyABCD were detected in only 53% of ST131 and 27% of ST69 isolates. The
cnf1 gene was detected in 53% of the ST131 isolates. The presence of these genes in ST131 isolates suggests that they exhibit high virulence and toxicity compared with non-ST131 isolates. The IbeA protein recognizes surface receptors on brain capillary endothelial cells, allowing the pathogen to invade the nervous system and cause neonatal meningitis [
21–
23]. Therefore,
ibeA is a representative VF gene in NMEC strains. In our study, three of each of ST357, ST372, and ST998 isolates (5%) carried
ibeA. Major UTI pathogens, such as ST131 and ST1193, may not carry
ibeA and therefore, do not cause meningitis in neonates.
ST131 isolates carried more VF genes, including
tsh and
espC, than non-ST131 isolates. The VF genes in ST131 isolates may play a role in pathogenesis. In a previous study,
tsh was present in >50% of avian pathogenic
E. coli isolates, 4.5% of UPEC isolates, and 11.5% of NMEC isolates [
15]. In this study, all ST131 and ST1193 isolates among UTI-associated
E. coli isolates carried
tsh. Although the function of the temperature-sensitive hemagglutinin encoded by
tsh in human infections is unclear, in Bacteroides fragilis, this protein contributes to abscess formation in intra-abdominal infections [
24]. ST127 isolates can cause bacteremia in adults and are highly virulent in experimental models of invasive
E. coli infection. Therefore, ST127 isolates should be monitored because of their greater pathogenic potential than more widely recognized clones, including ST73, ST95, and ST131 [
25]. Two ST127 isolates were recovered from urine in this study and had high virulence scores of 20 and 17.
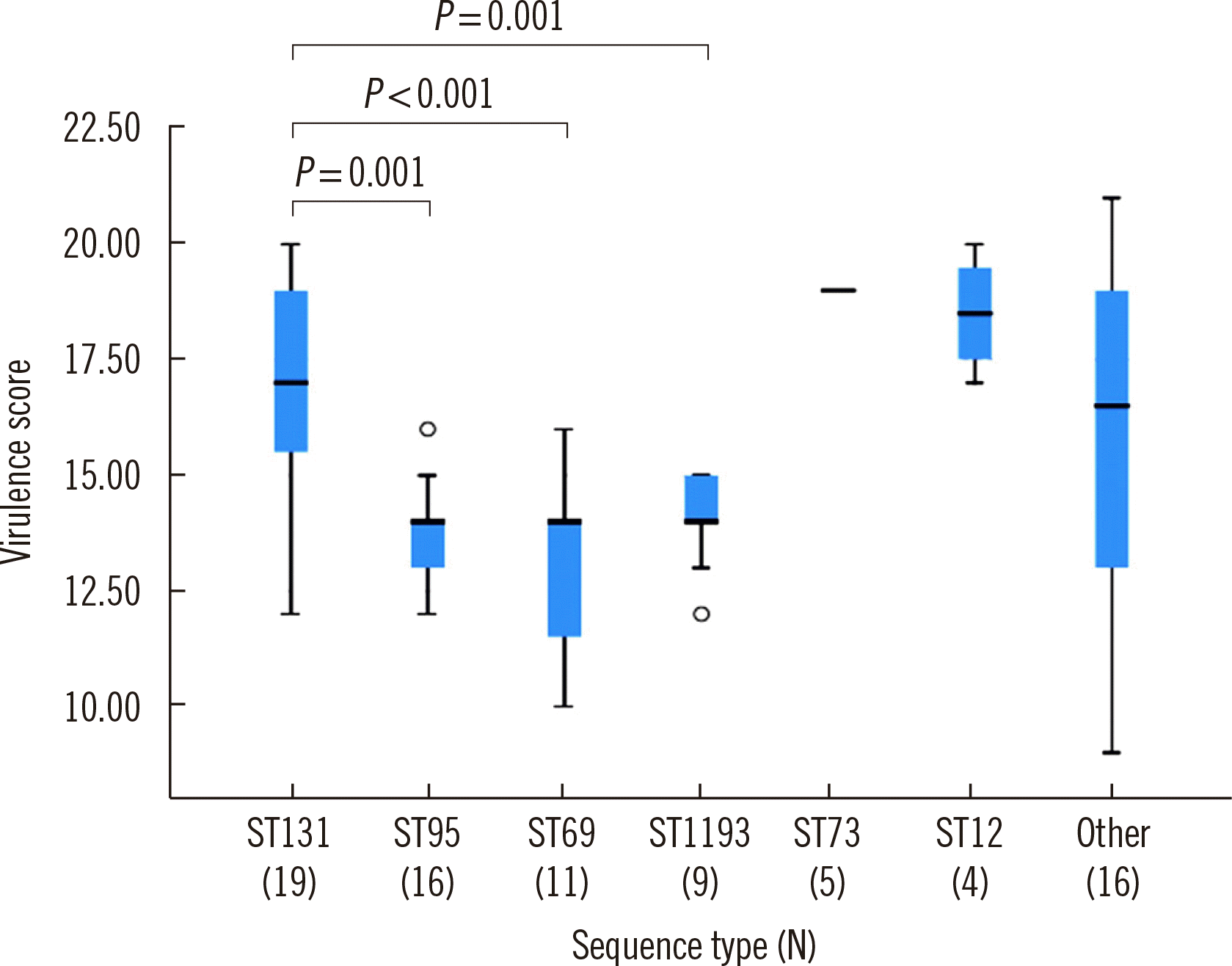
ST131 isolates reportedly had a higher virulence potential than other
E. coli types in a murine sepsis model [
5]. Similarly, in this study, ST131 isolates had higher virulence scores than ST1193, ST95, and ST69 isolates (
P<0.05). Shah,
et al. [
26] suggested a significant association between VFs and antimicrobial resistance in UPEC. Consistent herewith, we found that ST131 isolates of UTI patients had several VFs and a high resistance rate to several antibiotics.
Our study had some limitations, mainly related to the relatively small specimen size. Because of the limited number of E. coli bacteremia isolates reported, larger studies are needed to accurately determine the VF composition of these isolates in relation to their clonal characteristics. Further studies are needed to clarify the association between virulence and antibiotic resistance in urosepsis-related E. coli strains. Second, WGS analysis and web-based assessment are subject to experimental error. Third, the association between VFs and disease severity could not be clarified in this study.
In conclusion, E. coli bacteremia isolates from UTI patients were analyzed for a broad range of VFs, molecular backgrounds, and antibiotic resistance genes using WGS. We found no STs and VFs associated with bacteremia in WGS data of E. coli isolated from UTI patients. ST131, a pandemic multidrug-resistant clone present in both blood and urine specimens, was the most frequent ST and carried more VF genes, especially tsh and espC, than non-ST131 isolates.
Go to :





 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download