INTRODUCTION
Healthcare-associated infection (HAI) includes nosocomial infections occurring in hospitals, out-of-hospital infections resulting from discharge, and outbreaks in the community (1). According to a study in the United States, HAI caused by multidrug-resistant bacteria infections occur in more than two million people each year. It was confirmed to account for 1% of all deaths (2). HAI includes surgical wound infections, bloodstream infections, urinary tract infections, and respiratory infections. Patients with significantly reduced immunity, such as the elderly, chronically ill, and organ transplant patients, are particularly vulnerable to HAI. Among the various treatments of patients, renal patients undergoing hemodialysis are treated with reduced immunity and are easily exposed to HAI, including injection needle accidents and multidrug-resistant bacteria, so infection control for hemodialysis is important (3, 4). According to the Korean Nephrological Association’s report “2018 Korea’s Renal Replacement Therapy,” the number of patients undergoing renal replacement therapy in Korea is 98,746, of whom 73,059 are undergoing hemodialysis. In particular, the number of renal failure patients per one million population was reported as 1,907. The number of people with renal failure is increasing every year, and the mortality rate is high, so many academics and the media are interested. Chronic kidney disease is generally associated with old age, diabetes, hypertension, obesity, and cardiovascular disease, and hemodialysis is a major treatment for these kidney diseases (5). Most patients with CKD receive hemodialysis three times a week, and the dialysis regimen is targeted to eliminate about two-thirds of urea during each treatment (6). Waste products, such as urea, diffuse through a thin membrane that separates the dialysate flowing in the opposite direction from the blood. Most bacteria and viruses present in dialysate have a high molecular weight and cannot pass through a thin membrane. However, toxins and the like have a small molecular weight, so they can pass through the membrane, requiring special attention in the use of the dialysate (7). The quality management of dialysis water used as dialysate is important for patients exposed to large amounts of water (8). Abnormalities in the water purification facility of dialysis water cause chemical and microbiological contamination, and dialysis water contaminated with bacteria causes not only inflammatory reactions but also various diseases due to the influx of toxins into the body (9). Fever from bacterial infection is caused by bacterial inflammatory substances and stimulates the secretion of interleukin-1 (IL-1) and cytokines by peripheral blood mononuclear cells (PBMC) (10). IL-1 causes an increase in C-reactive protein (CRP), and cytokine mediates the host response to infection and is involved in acute and chronic inflammation of bacterial ions (11). Therefore, even if there is no sign of fever, increased cytokine production due to the contamination of dialysis water causes chronic inflammatory conditions, resulting in dialysis-related amyloidosis, hypoalbuminemia, atherosclerosis, hypotension, and coma (12). In medical institutions that perform dialysis, HAI should be prevented through the regular monitoring of dialysis water. Detailed guidelines for managing dialysis water are provided by organizations such as the Association for the Advancement of Medical Instrumentation (AAMI), the American National Standards Institute (ANSI), the International Organization for Standardization (ISO), and the European Best Practice Guidelines (EBPG). According to the AAMI guidelines (13), the quality control of dialysis water includes a chemical test (Table 1) to confirm the presence of heavy metals and a microbiological test (Table 2) to confirm the presence or absence of endotoxins and microorganisms. According to the AAMI guidelines, the acceptance criteria for dialysis water are less than 100 CFU/ml of bacteria and less than 0.25 EU/ml of endotoxins. In dialysis water, the action level (the concentration of bacterial contamination that must be taken before the maximum limit) is 50 CFU/ml of bacteria and 0.125 EU/ml of endotoxins, and corrective action must be taken quickly to drop below this level. As for the enforcement standards for each test, chemical tests should be conducted at least once a year, endotoxin tests must be conducted once a quarter, and microbial culture tests must be conducted once a month. For microbial culture tests, Trypticase Soy Agar (TSA) is recommended by the AAMI in the United States, and Reasoner’s 2a Agar (R2A) is recommended by the International Organization for Standardization (ISO) (14). R2A and TSA studies reported that microbial cultures using R2A showed higher microbial detection compared to TSA cultures (12, 15). However, according to a study published in 2016, there was no significant distinction in microbial cultures using TSA and R2A (16). Looking at the above foreign cases, differences were confirmed in R2A and TSA. In addition, foreign research data did not include studies on environmental factors such as climate, season, and water quality in the country. In Korea, studies related to microbial culture tests and environmental factors for R2A and TSA in hemodialysis water are insufficient, and studies suitable for the domestic environment have been judged necessary to control infection and prevent HAI in hemodialysis water.
Table 1.
Maximum allowable levels of toxic chemicals and dialysis fluid electrolytes in dialysis water, Maximum allowable levels of trace elements in dialysis water*
Therefore, this study aims to analyze the microbial culture and sensitivity of R2A, and TSA used in microbiological tests of hemodialysis water, identify seasonal microorganisms, and use them as basic data for improving hemodialysis infection control.
Go to : 
MATERIALS AND METHODS
Study sample
The hemodialysis water used in this study was a sample requested for microbial culture from September 2017 to August 2018, and 35 samples per month, a total of 420 samples were studied. According to the type of specimen, 396 cases of dialysis water and 24 cases of reverse osmosis water are classified.
Sample Transport
The samples were collected in a sterile, endotoxin-free container, and immediately transported to the laboratory for testing within 30 minutes.
Inoculation Media
Specimens requested from the laboratory were inoculated evenly on the surface of the medium by the spread plate method using a pipette in a TSA (Micromedia Co. Ltd., Korea) and an R2A (Asan Co. Ltd., Korea) medium, respectively. All tests were conducted in a biological safety cabinet (BSC class II), and the same specimen was repeated three times.
Culture Methods
As the culture media, TSA (Micromedia Co. Ltd., Korea) and R2A (Asan Co. Ltd., Korea) were selected, and an oxygen culture environment was selected as the culture condition. Regarding the incubation period, the TSA medium was cultured at 37°C for 48 hours, and the R2A medium was cultured at 27°C for one week (Table 3).
Gram Stain
The pure cultured independent colonies were smeared on slides, dried, and stained according to the manufacturer’s standard usage guidelines using a Gram stain reagent (YD Diagnostics Co. Ltd., Korea). The staining and morphological characteristics of the microorganisms were observed using a microscope (x1,000). Microorganisms were identified by dividing them into gram-positive bacteria, gram-negative bacteria, and fungi through microscope checks.
Statistical Analysis
All experimental results were expressed as mean and standard deviation through a total of three experiments, and the statistical analysis tested for the mean value and significance using SPSS Version 25.0 for the window software program. The significance test was performed at the p < 0.05 level.
Go to : 
RESULTS
Comparing the number of dialysis water samples with bacterial culture by R2A and TSA
Of the total 420 hemodialysis water samples, 28 cases (6.7%) showed positive results for both R2A and TSA, and 71 cases (16.9%) were positive for R2A and negative for TSA. There were 19 samples (4.5%) that showed negative results in R2A and positive results in TSA, and 302 cases (71.9%) were confirmed as negative results in both R2A and TSA (Table 4, Fig. 1).
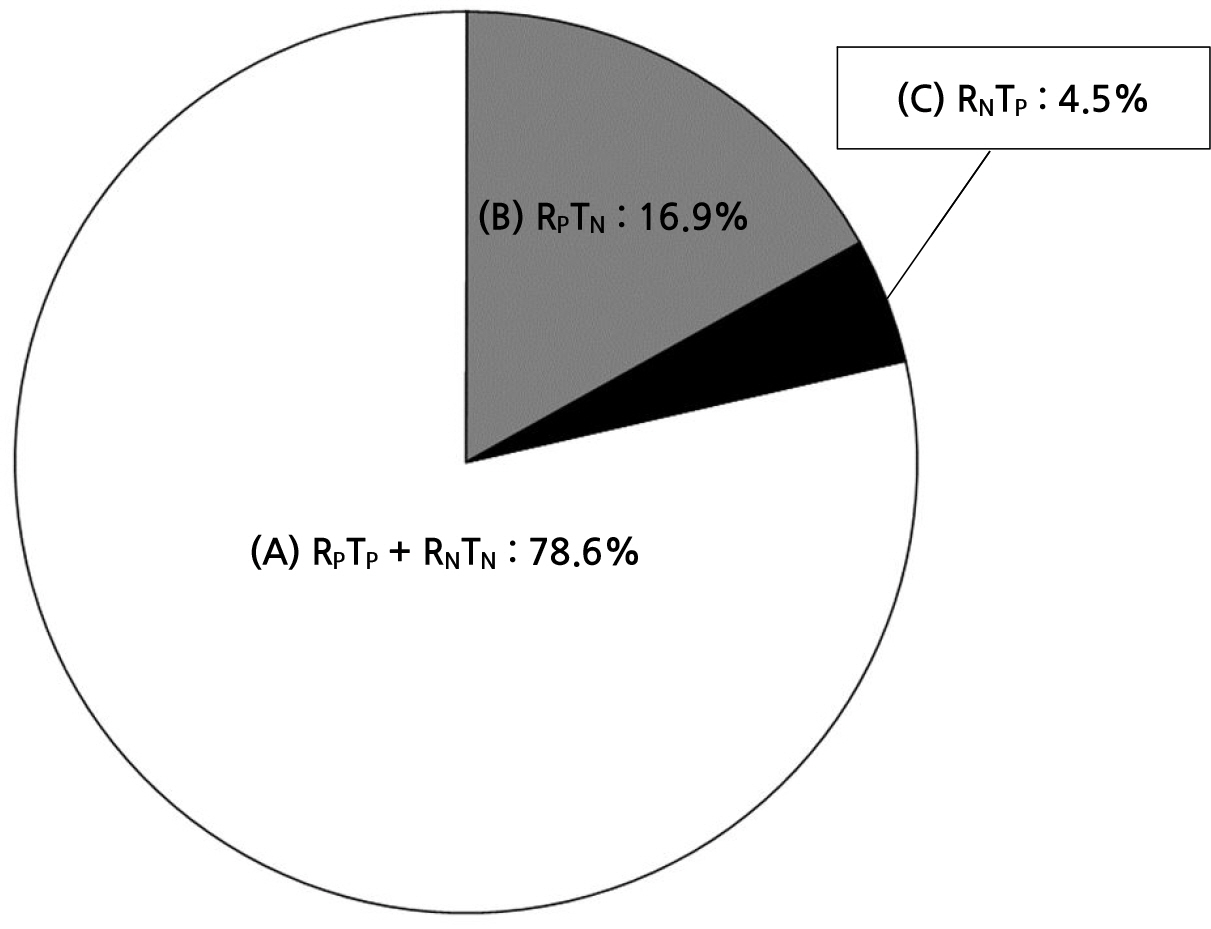 | Fig. 1Comparison of techniques for culturing dialysis water. (A) The positive growth of dialysis water and the negative growth of dialysis water were identified as 78.6% in both R2A and TSA. (B) The positive growth on R2A and negative growth of TSA were identified as 16.9% in dialysis water. (C) The positive growth on TSA and negative growth of R2A were identified as 4.5% in dialysis water. |
Table 4.
Comparing the number of dialysis water samples with bacterial culture by R2A and TSA
| Number of samples | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sep | Oct | Nov | Dec | Jan | Feb | Mar | Apr | May | Jun | Jul | Aug |
Total (%) |
|
| RPTP | 6 | 2 | 2 | 1 | 3 | 2 | 1 | 3 | 1 | 2 | 1 | 4 |
28 (6.7) |
| RPTN | 8 | 8 | 4 | 3 | 14 | 2 | 3 | 3 | 13 | 2 | 6 | 5 |
71 (16.9) |
| RNTP | 2 | 2 | 3 | 3 | 0 | 1 | 1 | 1 | 3 | 1 | 1 | 1 |
19 (4.5) |
| RNTN | 19 | 23 | 26 | 28 | 18 | 30 | 30 | 28 | 18 | 30 | 27 | 25 |
302 (71.9) |
| Total | 35 | 35 | 35 | 35 | 35 | 35 | 35 | 35 | 35 | 35 | 35 | 35 |
420 (100) |
Comparison of the number of samples of dialysis water with bacterial positive cultures by R2A and TSA
In confirming the positive rate of each medium in all samples, the microorganisms detected in R2A were higher than those detected in TSA, with 99 cases (23.5%) in R2A and 47 cases (11.1%) in TSA (Table 5).
Table 5.
Comparison of the number of samples of dialysis water with bacterial positive cultures by R2A and TSA
|
Medium (TN)* |
Number of samples | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sep | Oct | Nov | Dec | Jan | Feb | Mar | Apr | May | Jun | Jul | Aug |
Total (%) |
|
|
R2A (420) |
14 | 10 | 6 | 4 | 17 | 4 | 4 | 6 | 14 | 4 | 7 | 9 |
99 (23.5) |
|
TSA (420) |
8 | 4 | 5 | 4 | 3 | 3 | 2 | 4 | 4 | 3 | 2 | 5 |
47 (11.1) |
Comparison of bacteria growth on R2A and TSA media in identifying samples ≥50 CFU/ml
Among the 420 specimens, 12 (2.8%) in R2A and 18 (4.2%) in TSA were found to be cultured with more than 50 CFU/ml, and TSA was more sensitive than R2A (Table 6).
Table 6.
Comparison of bacteria growth on R2A and TSA media in identifying samples ≥50 CFU/ml
|
Medium (TN)* |
Number of samples | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sep | Oct | Nov | Dec | Jan | Feb | Mar | Apr | May | Jun | Jul | Aug |
Total (%) |
|
|
R2A (420) |
1 | 2 | 2 | 0 | 3 | 0 | 0 | 2 | 0 | 0 | 2 | 0 |
12 (2.8) |
|
TSA (420) |
3 | 2 | 3 | 0 | 1 | 1 | 0 | 4 | 2 | 2 | 0 | 0 |
18 (4.2) |
Comparison of bacteria growth on R2A and TSA media in identifying samples <50 CFU/ml
Among the 420 specimens, 87 (20.7%) in R2A and 29 (6.9%) in TSA were found to be cultured with less than 50 CFU/ml, and R2A was more sensitive than TSA (Table 7).
Table 7.
Comparison of bacteria growth on R2A and TSA media in identifying samples <50 CFU/ml
|
Medium (TN)* |
Number of samples | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sep | Oct | Nov | Dec | Jan | Feb | Mar | Apr | May | Jun | Jul | Aug |
Total (%) |
|
|
R2A (420) |
13 | 8 | 4 | 4 | 14 | 4 | 4 | 4 | 14 | 4 | 5 | 9 |
87 (20.7) |
|
TSA (420) |
5 | 2 | 2 | 4 | 2 | 2 | 2 | 0 | 2 | 1 | 2 | 5 |
29 (6.9) |
Identification of heterotrophic bacteria isolated from dialysis water from September 2017 to August 2018
Of the 30 strains isolated from a total of 420 specimens, 188 strains were subcultured and studied. The isolated strains for each medium were identified as 126 in R2A and 62 in TSA, and the morphological characteristics of the microorganisms were confirmed through Gram staining.
In R2A, 42 Gram-positive strains (33.3%), 71 Gram-negative strains (56.3%), and yeast-like fungi accounted for 13 strains (10.3%). In TSA, 21 Gram-positive strains (33.8%), 40 Gram-negative strains (64.5%), and yeast-like fungi accounted for one strain (1.6%; Table 8).
Table 8.
Identification of heterotrophic bacteria isolated from dialysis water from September 2017 to August 2018
Seasonal distribution of bacterial populations isolated from dialysis water
The species of microorganisms separated from hemodialysis water were analyzed seasonally according to environmental factors, such as season and climate, in Korea (Table 9).
Table 9.
Seasonal distribution of bacterial populations isolated from dialysis water
Seasonal comparison of the number of samples of dialysis water with bacterial positive culture by R2A and TSA
Because of the seasonal analysis of the one-year study, the positive cases of microorganisms isolated from R2A were confirmed to be 24 cases in spring, 19 cases in summer, 30 cases in autumn, and 26 cases in winter. The positive cases of microorganisms isolated from TSA were 10 cases in spring, nine cases in summer, 17 cases in autumn, and 11 cases in winter. It was confirmed that the positive rate of microorganisms cultured in R2A medium was higher in all four seasons than that of microorganisms cultivated in TSA, and a high positive rate was confirmed in autumn in both R2A and TSA mediums, showing characteristic results (Table 10).
Go to : 
DISCUSSION
The influx of microorganisms into the body is a risk factor that threatens human health by causing inflammatory reactions and various diseases. The rapid cultivation and identification of microorganisms is required for the treatment and prognosis of diseases. Consequently, various media have been developed, and there have been changes in the culture environment and conditions. In particular, the selection of the medium is the most important factor in increasing the detection rate of microorganisms. In this study, microbiological tests of hemodialysis water were performed using R2A and TSA to confirm the positive rate for each medium, the positive rate for each season, and the results of microbial identification. This contributed greatly to the prevention of medical-related infections and the management of artificial kidney center infections implemented in medical institutions. It is the first in the world to reflect climate change and seasonal specificity, and it is a meaningful study conducted over a long time period. The positive rates of microorganisms in hemodialysis water were confirmed in 99 cases (23.5%) in the R2A medium and 47 cases (11.1%) in the TSA medium. Therefore, the R2A medium was found to be more sensitive than the TSA medium. In the Netherlands (15), in a study conducted on 229 samples of hemodialysis water and reverse osmosis water, the R2A medium showed a higher positive rate than the TSA medium, confirming results similar to this study. In Thailand (12), a study conducted on 143 samples of reverse osmosis water, also confirmed that the R2A medium was more sensitive than the TSA medium. The positive rate of bacteria counts of 50 CFU/mL or higher was found to be 2.8% in R2A and 4.2% in TSA in this study, which was higher than that of 1.5% in R2A and 1.3% in TSA, the results of the US study. When comparing the results with those of advanced countries, it is thought that a more active management of hemodialysis water is necessary. R2A is a low-nutrient agar used with lower incubation temperatures and longer incubation times (17). However, TSA is a universal medium containing two peptones to support the growth of various microorganisms (18). Hence, the identification and positive rates of microorganisms cultured in the two media are considered different. The 188 strains of 30 species isolated in this study were classified into 33.5% Gram-positive microorganisms, 59.0% Gram-negative microorganisms, and 7.4% yeast-like fungi. Aeromonas sp., rarely cultured in TSA, is a bacterium that exists in fresh water and has been reported as a zoonotic pathogen that causes corneal inflammation in a recent domestic study (19). Moraxella sp. is also known as the causative agent of infectious diseases of otolaryngology (20). In Japan, bloodstream infections caused by Methylobacterium sp. have been reported in patients undergoing hemodialysis (21). Especially Sphingomonas paucimobilis, Acinetobacter lwoffii, and Oligella ureolytica showed high separation rates among Gram-negative microorganisms. In a Thai study, Pseudomonas spp. was 40%, Moraxella spp. 23%, Acinetobacter spp. 16%, Staphylococcus spp. 16%, Alcaligenase spp. 14%, Gram-negative rod 7%, Corynebacterium spp. 3%, Micrococcus spp. 3%, Bacillus spp. 1%, Chromobacterium spp. 1%, Gram-positive rod 1%, Rhodococcus spp. 1%, and Streptococcus spp. 1% was isolated, which was like this study in that the separation rate of Gram-negative microorganisms was high (12). The species of microorganisms isolated from hemodialysis water differed from season to season, and Acinetobacter lwoffii and Sphingomonas paucimobilis were identified as bacteria isolated throughout the year from R2A. Sphingomonas paucimobilis is an opportunistic pathogen that causes meningitis, sepsis, bacteremia, and peritonitis in people with reduced immunity (22). Acinetobacter lwoffii is a potential opportunistic pathogen in patients with impaired immune systems, and it has been identified as a cause of healthcare-associated infections (23). It is necessary to manage infections more effectively for hemodialysis water by monitoring bacteria that are separated year-round and bacteria that are separated by season. In foreign countries, there have been no studies on the detection of microorganisms in hemodialysis water related to season or climate. However, in Korea, the four seasons are distinct, and the temperature change varies greatly from season to season. The cultivation of microorganisms is closely related to the temperature and growth of bacteria, as well as the components of the medium and cultivation time. Therefore, if the R2A medium and the TSA medium are used together for the microbiological testing of hemodialysis water, it is believed that they can contribute to patient safety and public health safety infections.
Go to : 




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download