INTRODUCTION
Human immunodeficiency virus-1 (HIV-1) infection remains a major public health issue. The developed antiretroviral therapy (ART) prevents the progression of acquired immunodeficiency syndrome (AIDS). Despite the emergence of various effective therapies, the interruption of ART induces the viral rebound to pre-ART levels (Fig. 1) (1). It has been known that the main obstacle to eliminating HIV-1 may be caused by latent viral reservoirs which have been considered as cells harboring replication-competent HIV-1 resident in patients treated with ART. Nevertheless, it is less known that the characteristics of viral reservoirs regarding their dynamics and heterogeneous nature (2, 3, 4, 5, 6). The viral reservoir is generally constituted by different long-lived cell types that provide a replication-competent provirus (7). CD4 T cells associated with immunological memory likely play a main role in persistent HIV-1 infection due to longevity and renewal through homeostatic proliferation (4). Although HIV-1 DNA has been detected in a number of different CD4 T cell subsets, some part of them have been considered as reservoirs containing the replication competent HIV-1 under ART treatment (8, 9, 10, 11). Among the T cells classified according to their differentiation status, memory T cells including central memory (TCM) , transitional memory (TTM) and effector memory T cells (TEM) are known as a main subsets of reservoir for replication-competent HIV-1(4, 12). Because the reservoirs exhibit the memory T cell properties, they mainly hide at anatomical sites such as lymphoid organs. Thus, viruses propagated from reservoirs activated in lymphoid organs rapidly spread to entire body via blood stream or cell-to-cell transmission during interruption of ART (13).
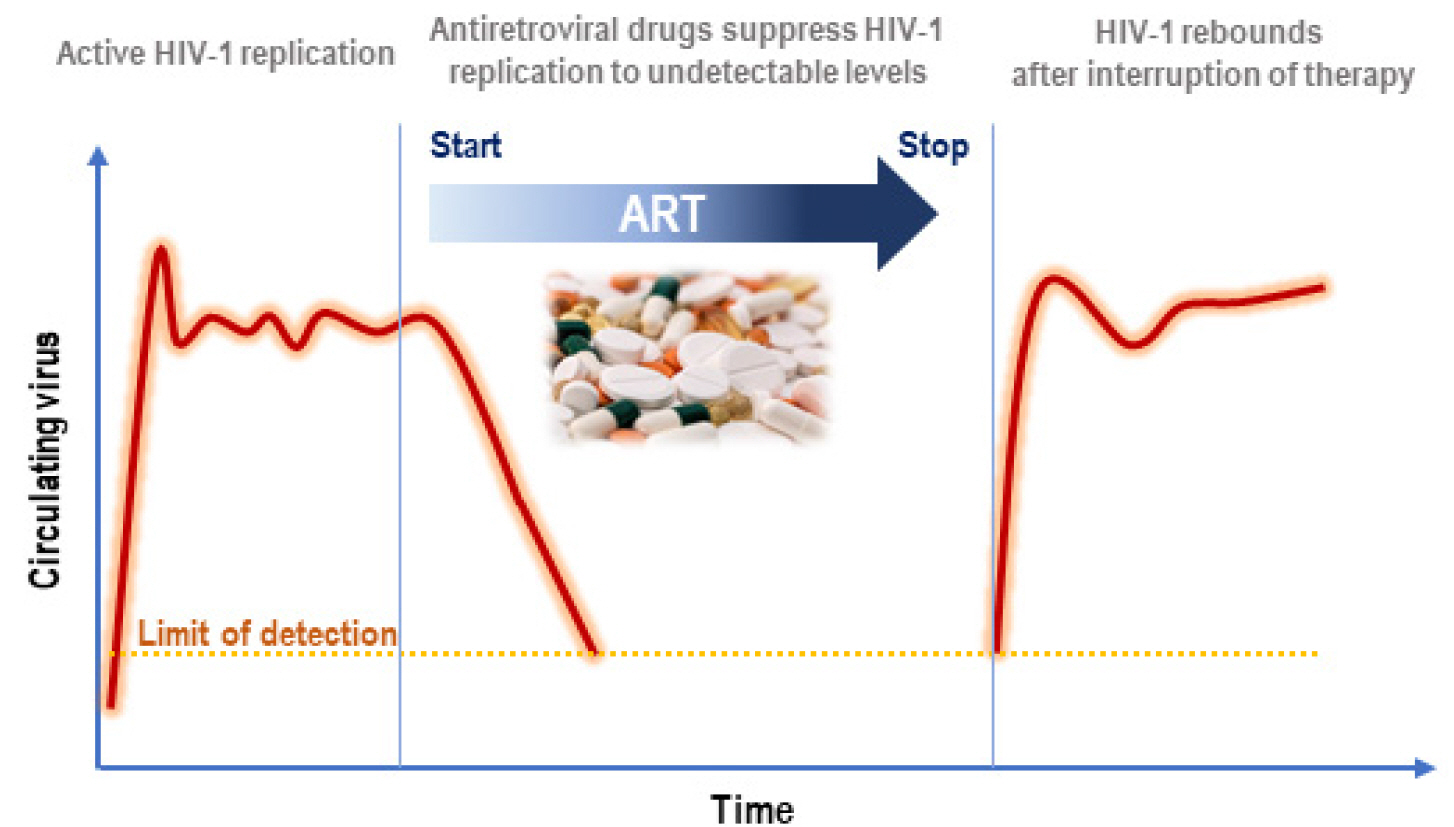 | Fig. 1Clinical definition of HIV-1 reservoirs (Adapted from Deanna A Kulpa et al., J Virus Erad. 2015 Apr; 1(2): 59–68.) (1). |
To attempt to eliminate the latent reservoirs, it is most important to determine the biomarker capable to probe the reservoirs. Thus we will overview the various biomarkers described in previous studies using latently HIV-1 infected cells and introduce ongoing research aimed at defining the latent reservoir at the Korea National Institute of Health (KNIH).
Go to : 
THE LATENCY OF HIV-1 IN T-CELL SUBSETS
As for therapeutic strategies aimed at eliminating latent HIV-1 reservoirs, it is important to understand the cell type harboring the latent HIV-1 provirus. Notably, the memory CD4 T cells known as a major reservoir, has various phenotypes according to their differentiation and memory status (9, 10, 11, 14). Fortunately, the developmental profile of memory CD4 T cell has been well conceptualized, and is best characterized as a differentiation linear model (2). HIV-1 can persist in naïve (TN), stem cell-like memory (Tscm), central (TCM), transitional (TTM), and effector (TEM) memory CD4 T cells with different infection rates. Even though the frequency of HIV-1-infected cells is lower in TN compared to TCM (TN: 684 vs TCM: 1,279 HIV-1 DNA copies/ml, respectively), Zerbato et al. showed that TN harbors highly inducible replication-competent provirus (TN: 18,290 vs TCM: 15,135 HIV-1 RNA copies/ml, respectively), indicating that TN might not play a role as reservoir (10). Hiener et al. demonstrated that the TEM of patients treated with ART contained the highest number of intact HIV-1 proviruses (14). These results provide the evidence which the frequency of intact HIV-1 proviruses differed across the T cell subtypes.
T helper type 1 cell (Th1), Th2, Th17, Th9, regulatory T cell (Treg), and follicular T helper cell (Tfh) are classified by functional polarization, and these characteristics affect the HIV-1 to be persisted (5, 15, 16). According to a recent study, proliferation of HIV-1-infected Th1 cells play a crucial role in maintaining the number of cells that harbor replication-competent HIV-1 (5). As observed in the digestive system, it was also found that T helper type 17 (Th17) cells contributed to persistence of HIV-1 in ART-treated individuals (6). Furthermore, CD4 T cells polarized to Th1/17 harbor high portion of HIV-1 DNA in ART treated individuals and contribute disproportionately to the viral reservoirs remained stably during ART (15).
These studies emphasize the heterogenous natures of HIV-1 reservoirs and stress the urgent need to identify markers for latent reservoirs. If reservoirs could be isolated using a useful marker, the markers may be helpful to defining the various and complex characteristics of the reservoirs and could even make possible selective targeting for the complete eradication of the virus. So far, no cellular marker capable of distinguishing all HIV-1 reservoirs has been found; however, some HIV-1 reservoir markers have been proposed recently, and their characterization will be an important contribution to our understanding of HIV-1 reservoirs.
Go to : 
IMMUNE CHECKPOINT MOLECULES (IC)
Chronic HIV-1 infection, with its high antigen load, constantly stimulates T cells, which then brings about their dysfunction, called “T-cell exhaustion” (17, 18). During this period, the expression of several immune checkpoint molecules (ICs) increases in the T cell, which suppress the immune response, thereby increasing potential HIV-1 latent infection. These ICs include T cell immunoglobulin and immuno-receptor tyrosine-based inhibitory motif (ITIM) domain (TIGIT), programmed cell death-1 (PD-1), lymphocyte activation gene 3 (LAG-3), cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), T cell immunoglobulin and mucin 3 (TIM-3), CD160, and 2B4 (CD244) (19, 20). Fromentin et al. demonstrated that TIGIT, PD-1, and LAG-3 were related to HIV-1 persistence in CD4+ T cells (19). Although differences exist depending on the subset of the memory CD4+ T cell, the majority of integrated HIV-1 DNA was found to be high in cells with at least one marker. In particular, it was observed that the level of integrated HIV-1 DNA was 8.2-fold higher in cells which expressed all three markers, compared with total CD4+ T cells (19) (Table 1). Recently, the same group reported that the ratio of HIV-1 p24 (capsid protein)-producing cells was higher in cells enriched as a single antibody panel with TIGIT, PD-1, and α4β1 (median fold 2.23, 1.65 and 2.21, respectively) using samples taken from ART patients (20).
Table 1.
The surface molecules reported as biomarkers targeting HIV-1 reservoirs
| Marker |
ATR Median time |
Cell Type | Outcome | Characterization & Relevance | Reference |
|---|---|---|---|---|---|
|
PD-1 TIGIT LAG-3 |
8.5 years | Memory CD4+ T cell | Integrated HIV DNA | ·PD-1+TIGIT+LAG-3+CD4+ T cell/ total CD4+ T cell = 8.2-fold | (19, 20) |
| TILDA | ·PD-1+, TIGIT+ or LAG-3+ CD4+ T cell/ total memory CD4+ T cell : 76% inducible reservoir | ||||
| CD32ahigh | 34 month | CD4+ T cell | HIV DNA |
·CD32ahi / total CD4+ T cells = 633-fold ·CD32ahi / CD32a-CD4+ T cells = 1024-fold ·Population CD32a+ / total CD4+ T cell = 26.8~86.3 % |
(37) |
| qVOA | ·CD32a+CD4 T cell / total CD4+ T cell = 3000 | ||||
| CD30 | ART patients | CD4+ T cell | HIV RNA |
·Most CD30+CD4+ T cells : effector/transitional memory phenotype ·HIV RNA from CD30+CD4+ T cells = 21% ·Population CD30+CD4+ T cells / total CD4+ T cell = 3.97 % |
(28) |
| CD2high | < 3years | Resting memory CD4+ T cell | HIV DNA | ·CD2hi CD4+/ total CD4+ T cells = 5.7 fold | (32) |
| CD20dim | 28 month | Resting CD4+ T cell |
HIV RNA HIV DNA |
·CD20dim CD4+ T cells / total pool HIV-expressing cells = 18.55% ·Resting CD20dim CD4+ T cells / total CD4+ T cells = 2.1 fold |
(34) |
The Perreau group reported that PD-1+/Tfh cells containing replication-competent virus in lymph node were detected highly in ART-treated aviremic HIV-1-infected individuals (21). In the follow-up study, the ratio of CXC chemokine receptor 3+ (CXCR3) was found to be higher in PD-1+/Tfh memory CD4 cells (22).
IC inhibitors have already been used in cancer treatment and even been suggested as possible agents for use in HIV-1 infection cure, but their application as a treatment agent has limitations due to the inconsistent results of related clinical trials and their toxicity.
CD30
CD30, a member of TNF receptor superfamily, is expressed in tumor cells such as Hodgkin's disease or aggressive lymphoma (23, 24). In particular, the rate of CD30 expression increases rapidly due to several viruses such as human T-cell lymphotropic virus and Epstein-Barr virus (25, 26). Although it has been known for twenty years that CD30 plays an important role in HIV-1 replication and HIV-1-infected CD4 T cell death (27), it was recently discovered that HIV-1 RNA is abundantly enriched in CD30+CD4+ T cells (28). In an in situ hybridization of gut-associated lymphoid tissues, the co-localization of CD30 and HIV-1 RNA was observed at 88% in the ART group and 32.5% in the viremic group. Also, after ex-vivo treatment with FDA-approved drugs (brentuximab vedotin, anti-CD30 antibody-conjugated drug) targeting CD30 in the peripheral blood mononuclear cell (PBMC) of patients, the total HIV-1 DNA was observed to decrease greatly (28).
Even though CD30 has been considered as a marker for HIV-1 replication rather than a viral persistence marker (28, 29, 30), it will be necessary to further investigate the basis for using it as an HIV-1 reservoir marker and to evaluate its possibility as a therapeutic target in a follow-up study due to lack of in vivo efficacy test yet.
CD2
CD2, a member of the immunoglobulin superfamily, is a specific marker for T cells and NK cells. Iglesias-Ussel et al. produced latently HIV-1 infected cells using an in-vitro primary CD4+ T cell model (31), and then their transcriptional activity was analyzed using the micro-array analysis. Twenty transcripts encoding cell surface proteins were up-regulated in comparison to these of uninfected cells. Of these, CD2 showed significant difference was further analyzed in patients’ samples (32). The ratio of integrated HIV-1 DNA was found to be higher in CD2high than in CD2low of the resting memory CD4 T cell (CD3+CD4+HLA-DR+CD45RA-) in the samples of ART patients, showing an increase of 5.7-fold compared to the total CD4 T cells (Table 1). In addition, viral RNA production increased in CD2high compared to CD2low, suggesting its potential as a new marker. Since it remains difficult to completely capture the latent infection reservoir, the relevance of CD2 still has to be studied in lymphoid tissues.
CD20
Although CD20 has been known as a B lymphocyte antigen initially, it is dimly expressed by CD4+ T cells of small population (1-2%) (33). Serra-Peinado et al. recently reported CD20 as a marker for HIV-1-infected CD4+ T cell. Although CD20-expressed CD4 T cells were rare, the number of cells expressed with CD20 was higher in infected cells compared to non-infected cells (34). These infected CD20+CD4+ T cells contributed medians of 18% and 25% to the total pool of HIV-1 RNA+ cells in ART-suppressed and viremic patients, respectively. Also, resting CD20+CD4+ T cells had higher levels of HIV DNA compared with the total CD4+ T cells (2.1-fold) (Table 1). In addition, ex-vivo treatment of PBMCs from ART-suppressed individuals with the anti-CD20 monoclonal antibody Rituximab appeared to greatly reduce the HIV-1 RNA+ pool. Rituximab deplete viral-reactivated cells, but it will be necessary to conduct a further study to evaluate the possible application of Rituximab as an appropriate treatment for latent infection by reducing the intact HIV DNA reservoir.
CD32a
CD32a, known as Fc gamma receptor IIa (FcγIIa), is expressed at a high level mainly in myeloid cells, and is expressed in a low rate in T cells (35, 36). Recently, Descours et al. discovered CD32a as a new marker for HIV-1 reservoir cells (37). In this paper, HIV-1 DNA was enriched by more than 1000 times with CD4+CD32a+ cells compared to CD4+CD32a- cells (Table 1). Accordingly, they demonstrated that the replication-competent provirus was enriched in CD4+CD32a+ cells. However, in the follow-up studies conducted by other groups, it was questioned whether CD32a was a real marker for HIV-1 latent reservoirs. In several papers, total HIV-1 DNA or replication-competent provirus were not enriched in CD32+ cells (38, 39, 40, 41). The main reason for such difficult reproducibility is the technical issue that isolates CD32+CD4+ T cells with high purity. CD32+ is expressed at a much higher rate in antigen-presenting cells (APCs) compared to CD4 T cells, and if the residual APCs are not separated and left, an excessive imbalance can develop as a result (42, 43). Also, CD32b, an isoform of CD32, is highly expressed in B cells, and no antibody targeting CD32a only is present as yet; thus the technology is important for finely separating T or B cells mixed blood cells using flow cytometry (42). Very recently, Darcis et al. proved the importance of purity at the level of CD4+ T cell isolation. The results of their study showed that the interference of CD4- T cells on CD32+ enrichment decreased as purity increased (42). Nevertheless, an additional study is needed to investigate the characterization of CD32+CD4+ cells in combination with other markers or the downstream of CD32a signaling because CD32a is related to HIV-1 infection.
CD127
Tissue memory CD4+ T cells (hereafter referred to as Tm cells) expressing IL-7 receptor-alpha (CD127) were suggested as a novel marker for latent HIV-1 reservoir (44). Previous study reported that CD127+ Tm cells were lack of productive HIV-1 infection (45). Hsiao et al. demonstrated that the level of integrated HIV-1 DNA was higher in CD127+ than CD127- Tm cells when sorted from patient’s tonsil were cultured by HLAC (human lymphoid aggregative culture) (44). Also, CD127+ Tm cells were supported low levels of productive infection compare to CD127- Tm cells. According to RNA-seq profiling, CD127+ Tm cells down-regulated gene pathways associated with active transcription of HIV-l, including NFκB and NFAT signaling. The study identified a novel subset of CD4+ T cells resident in tissues excepting blood that could be preferentially targeted for eliminating latent HIV-1 reservoir.
Go to : 
HIV-1 LATENT INFECTION STUDY AT THE KNIH
At the Korea National Institute of Health, studies have been conducted to discover the factors involved in the formation and maintenance of HIV-1 infection, in the form of latent infection research mainly through epigenetics. In particular, it is thought highly possible that genes for histone modifications which commonly change in several types of latent infection cell lines, play a crucial role in HIV-1 latent infection, and a subsequent follow-up study was performed to find out that the cell-cycle regulatory genes cyclin-dependent kinase inhibitor 1 (CDKN1A) and cyclin D2 played an important role in the maintenance of HIV-1 latent infection (46). Also, a study on the distribution of histone modifications by chromosome was performed to find out that a specific gene called nuclear factor 1 X type (NFIX) acted on the latent infection (47) (Fig. 2).
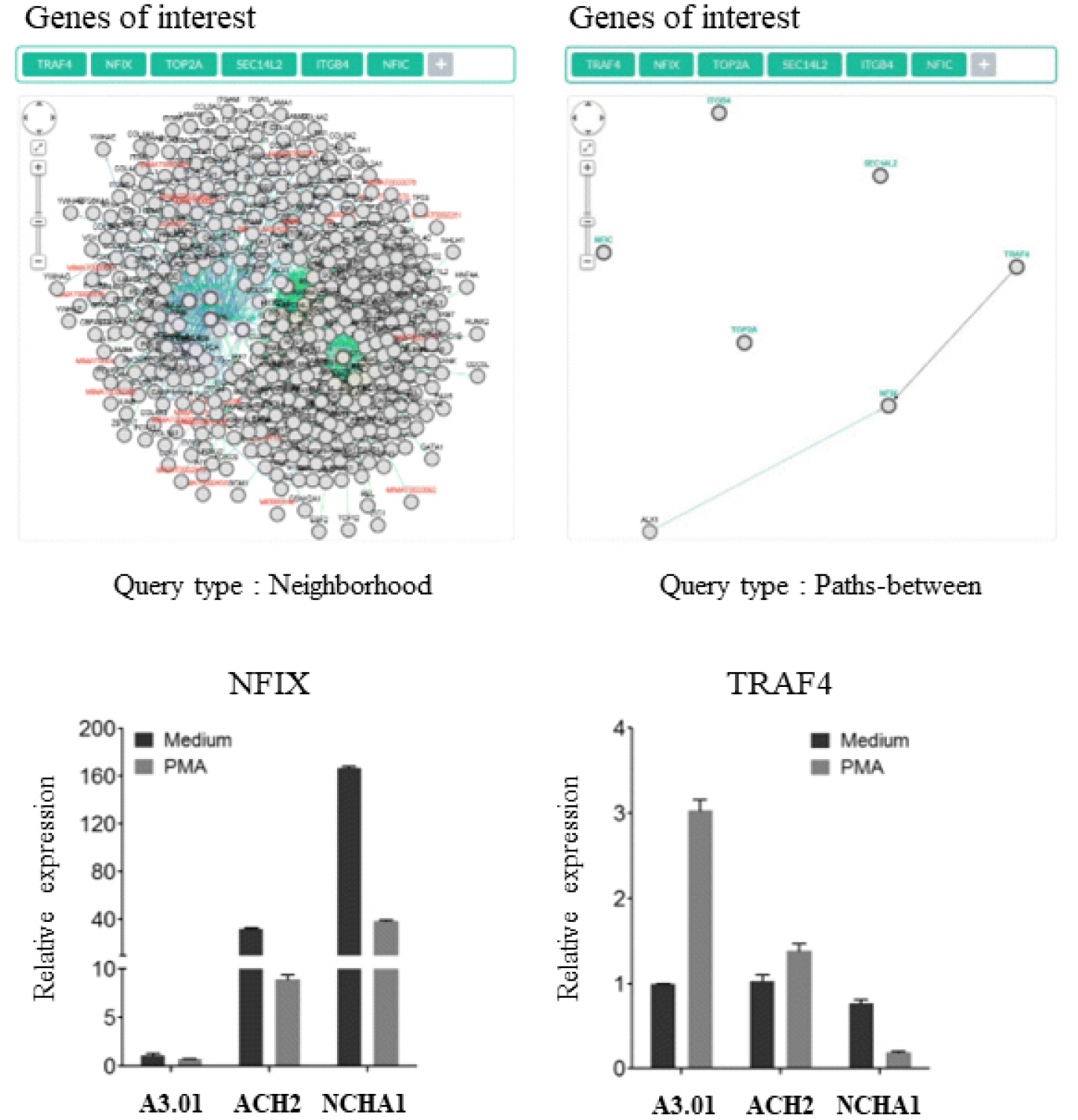 | Fig. 2Interaction of NFIX and TRAF4 in latently HIV-1 infected cell line (47). The expression of NFIX and TRAF4 is examined using real-time PCR between HIV-1 latently infected cell lines (ACH2 and NCHA1, and an uninfected cell line, A3.01). NFIX is highly expressed in ACH2 and NCAHA1 cell lines. |
In addition, studies aimed at discovering useful HIV-1 latent infection regulatory factors and their mechanisms and finding biomarkers for detecting latent infected cells are being performed using the PBMCs from patients. The PBMCs of long-term non-progressors (LTNPs) are obtained from patients already collected and stored at the College of Medicine of the University of Ulsan, an external joint researcher, which are then subjected to RNA-Seq technique and bioinformatics integrated analysis in order to find latent infection regulatory factors and to suggest them as biomarkers for detecting latent infection (48).
Go to : 
CLOSING REMARKS
HIV-1 reservoirs exist in various ways according to heterogenous localization, longevity, and activation status (3, 4, 5, 9, 10, 11, 14, 49, 50). It is therefore reasonable to assume that various cellular proteins are highly expressed in the reservoir cell. There are conflicting views on newly suggested markers, and contradictory results have been obtained depending on the various technological issues and experimental systems, infection and preparation processes, and methods of purifying patients' cells (37, 38, 39, 41, 43, 51, 52, 53, 54). According to studies on recently suggested CD32a in particular, the process of isolating cells is technically important (42). Nevertheless, it was demonstrated that the expression rates of HIV-1 RNA and DNA were high in CD32a+PD-1+CD4+ T cells or CD32+CD30+ cells (28, 55). At present, it is difficult to perfectly target an HIV-1 reservoir with a single surface marker, but it will eventually be possible to separate HIV-1 reservoirs with a higher rate through “multicolor” combinations of existing markers.
Finally, to understand the physiopathology of HIV-1 infection and establish appropriate treatment strategies, it is important to continue investigating the characteristics of the various T-cell subsets and non-lymphoid cells that are related to viral persistence and reactivation.
Go to : 




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download