Abstract
Background
Acute lymphoblastic leukemia (ALL) is the most common type of leukemia in children. Several environmental and genetic factors are known to be involved in its development and progression. The angiopoietin-Tie system is one of the most critical factors in angiogenesis, and its possible role in solid tumors and leukemia has been previously investigated. In this study, we examined the expression of these genes in ALL patients (early pre-B-ALL and pre-B-ALL) and compared them with normal samples.
Methods
Bone marrow samples were collected from 40 patients (aged 0‒19 yr) newly diagnosed with early pre-B-ALL or pre-B-ALL using molecular and flow cytometric tests and from 15 control individuals. For molecular tests, RNA extraction and cDNA synthesis were performed, and Ang1, Ang2, Ang4, Tie1, and Tie2 gene expression was examined by real-time polymerase chain reaction.
Leukemia is caused by the unrestrained proliferation of hematopoietic stem cells and was reported to account for 3.8% of cancer deaths as of 2020 [1]. Acute lymphoblastic leukemia (ALL) is a clonal malignancy of the lymphoid line of blood cells. It is the most common childhood cancer and the most common cause of cancer death in individuals younger than 20 years in the United States [2]. Based on the involved cell line, the World Health Organization divides ALL into B-ALL and T-ALL, each of which can be detected by immunophenotyping. B-ALL is the most prevalent type among both children (88%) and adults (75%) [3]. Despite numerous studies on the etiology of ALL, its causative agents have not been precisely identified. However, heredity, infections, and environmental factors such as radiation exposure play an essential role in ALL pathogenesis [2, 3]. Numerous studies have shown angiogenesis in leukemia by reporting increased levels of various angiogenic factors in patients and have suggested that anti-angiogenic drugs can act as a treatment agent for leukemia [4, 5]. The balance between angiogenic and anti-angiogenic factors secreted by vascular endothelial cells regulates angiogenesis, thus determining leukemia progression. Different types of angiogenic factors and their role in angiogenesis have been previously described; these factors include VEGF, FGF, angiopoietin, TGF, PIGF, HGF, HIF, and TNF [4, 6, 7].
The angiopoietin family (Ang1, Ang2, and Ang4) and its receptors (Tie1 and Tie2) are some of the most significant and well-known angiogenic factors [8]. Ang1 and Ang2 play antithetical roles in the process of angiogenesis, and Ang4, the human ortholog of Ang3, has a similar role to Ang1 [9, 10]. Ang1, one of the most noteworthy members of the Ang-Tie system, is a Tie2 agonist. By binding to and phosphorylating Tie2, this protein induces survival, proliferation, and migration signals in endothelial cells and, subsequently, maturation. It is also essential in inflammation [9, 11-13]. Ang2 acts as a natural Ang1-Tie2 antagonist. In the presence of VEGF, Ang2 causes the separation of pericytes from arteries. It also increases vascular permeability and prepares endothelial substrates for angiogenesis [10]. However, Ang2 can act as a relative Ang1 agonist with anti-apoptotic effects in cases of decreased Ang1 levels or high concentrations of Ang2 in vitro [14]. Ang4, much like Ang1, phosphorylates and activates Tie2 and its downstream pathways such as the PI3K signaling pathway. Furthermore, it causes endothelial cell survival, migration, and angiogenesis and increases blood vessel permeability. Ang4 also dilates lymph vessels and exhibits increased levels under hypoxic conditions in vitro [15]. However, little research has been done on this member of the angiopoietin family.
The primary receptor of angiopoietins is Tie2, which upon binding with ligands, is activated by autophosphorylation and activates its downstream pathways [16, 17]. A soluble type of the Tie2 protein regulates angiopoietin activity by binding to one angiopoietin type (most commonly, Ang1) to increase the activity of the other angiopoietins [18]. Tie1 is another angiopoietin receptor. No ligand is known for this receptor, and its function is induced through the formation of heterodimers with Tie2. Tie1 binds to Tie2 at the cell surface and prevents angiopoietins from binding to Tie2 [17]. The Ang-Tie system plays a key role in the angiogenesis, proliferation, and maturation of vascular endothelial cells and is a useful therapeutic target in solid tumors and hematologic malignancies [19]. However, few studies have been performed to determine expression level changes and the function of the angiopoietin family in ALL [20, 21].
In this study, we aimed to quantify the expression levels of the genes in the Ang-Tie system (Ang1, Ang2, Ang4, Tie1, and Tie2) in newly diagnosed ALL patients compared with those in normal samples.
Forty bone marrow (BM) samples from newly diagnosed ALL patients (aged 0–19 yr) and 15 normal BM samples (from individuals aged 0–19 yr) were obtained from the bone marrow transplantation section of Taleghani Hospital in Tehran from March to October 2020. Pre-B-ALL and early pre-B-ALL cases, which are the most common types of ALL, were included in this study. Normal BM specimens were selected from individuals who were referred to our hospital because of high peripheral blood (PB) WBC count, but their BM specimen examination and flow cytometric tests did not confirm any malignancy or other diseases influencing the BM. The patients signed a consent form, and questionnaires were filled out for demographic data collection. The study protocol was approved by the ethical committee of Tabriz University of Medical Sciences (code: IR.TBZMED.REC.1397.1040).
The nucleotide sequences of designed primers for the target and normalizer genes are shown in Table 1. Total RNA was extracted and purified using the Qiagen RNeasy Mini Kit (Cat. No: ID: 74104; Qiagen, Germany) according to the manufacturer's protocol. Quality and concentration assessments of the RNA samples were carried out using a Thermo Scientific NanoDrop One (Thermo Fisher Scientific Inc., Waltham, MA, USA). The Thermo Scientific SuperScript IV Reverse Transcriptase kit was used for cDNA synthesis. We chose the ABL gene as a normalizer gene, and relative expression of the Ang1, Ang2, Ang4, Tie1, and Tie2 genes were measured in BM samples of ALL patients and in normal samples. A total reaction volume of 15 mL was used for quantitative real-time polymerase chain reaction (qPCR). We used 7.5 mL of RealQ Plus Master Mix Green, without Rox (AMPLICON, Odense, Denmark) in each reaction, which was performed in the ABI StepOne Plus system in duplicates. As the first step of the qPCR, initial activation was performed at 95°C for 10 min. The thermal cycling program was carried out for 40 cycles with the following: denaturation (95°C for 10 s), annealing (58–64°C depending on the gene for 20 s), and extension (72°C for 30 s). After cycling, the final extension step (72°C for 10 min) was performed.
Real-time PCR efficiencies and the cycle threshold (CT) were used in the ^^CT formula [Δct (treated sample)–Δct (untreated sample)] to calculate the relative expression levels of the Ang1, Ang2, Ang4, Tie1, and Tie2 genes in ALL patients and normal samples. Data normalization of gene expression levels was performed using the ABL gene as a housekeeping gene. All statistical analyses were performed using Prism software version 8 (GraphPad, San Diego, CA, USA) and the SPSS statistical software package (version 18.0). The Kolmogorov–Smirnov test and the Mann-Whitney U test were used to check data normalization and to measure the differential expression levels of targeted genes in each sample type, respectively. The association between relative gene expression and demographic data was evaluated by Pearson's chi-square and Kruskal-Wallis tests. The diagnostic power of the ANG-TIE system genes was evaluated using receiver operating characteristic (ROC) curve analysis. Statistical significance was set at P<0.05.
In this study, 40 samples from newly diagnosed ALL patients and 15 samples from control individuals were collected. Demographic characteristics and data obtained from laboratory diagnoses of patients were collected using diagnostic tests or questionnaires filled out by the patients. Demographic and laboratory information of patients are shown in Table 2.
Statistical analyses were performed using the ABL gene as a housekeeping gene and 2^(–ΔΔct) to compare angiopoietin gene expression levels in ALL patients with control samples. Ang1 showed a significant decrease in expression levels in patients with ALL (fold change=0.3, P=0.0165) (Fig. 1).
Conversely, Ang2 (fold change=3.1, P≤0.0001), Tie1 (fold change=2.6, P=0.0014), and Tie2 (fold change=2.8, P=0.0001) were significantly upregulated in ALL samples compared to control samples (Fig. 1). However, Ang4 did not show a significant difference in expression levels between the two sample groups (P=0.09, Fig. 1).
We performed a correlation analysis between the expression levels of Ang-Tie system genes and ALL subtypes, patient age, blast count, sex, and PB WBC count. Table 3 shows that Ang-Tie system gene expression levels did not show a significant statistical associations across either patient demographic or clinical data.
ROC curves were plotted to evaluate the diagnostic roles of Ang1, Ang2, Ang4, Tie1, and Tie2 in ALL. Except for Ang4, the Ang-Tie system genes appeared to be appropriate diagnostic markers for ALL because their specificity, sensitivity, and the area under curve (AUC) values were higher than 70. In addition, the AUC values were statistically significant (P<0.05). Table 4 illustrates detailed information on the ROC curve analysis, and Fig. 2 depicts the diagnostic power of the genes.
ALL is caused by excessive proliferation and lack of differentiation in lymphoid blast cells, which eventually overwhelm the BM and PB. It is the most common cause of cancer-related deaths in individuals younger than 20 years in the United States [2].
Angiogenesis and its causative factors play critical roles in different types of cancers. In solid tumors, hypoxic conditions follow tumorigenesis, and the balance between pro-angiogenic and anti-angiogenic factors tends to tilt towards pro-angiogenic factors, which cause angiogenesis and tumor spread [4, 22]. Angiogenesis is also responsible for the spread of leukemia, and several studies have been performed to discern the effects of angiogenic factors in various forms of leukemia [4, 5]. The angiopoietin family and their receptors are among the most fundamental angiogenic factors [9, 10]. In the present study, we investigated the expression of genes belonging to this family, as well as of their receptors, in patients with ALL.
Our results indicated that Ang1 was expressed at lower levels in ALL patients than in control samples. The expression level of this gene was not significantly different across demographic or clinical characteristics (Table 3). Ang1 competes with Ang2 for binding to the Tie2 receptor, with Ang1 having a higher affinity. The expression of Ang1 is likely decreased in patients with ALL, leading to Ang2 activation. Examinations of Ang1 expression levels have shown variable results in different studies. In a study by Schliemann et al. [23], the expression of this gene in BM samples from AML (acute myeloid leukemia) patients did not show a statistically significant difference compared to normal samples. Conversely, other studies have shown Ang1 upregulation in AML patients, either in BM or PB samples [24-29]. In a study by Cheng et al. [28], increased expression of Ang1 was reported in patients with myelodysplastic syndrome (MDS). They also demonstrated an association between high Ang1 expression and short survival in patients with MDS [28]. On the other hand, Atesoglu et al. [30] and Karakurt et al. [20] demonstrated that the expression of Ang1 was reduced in AML and ALL patients, similar to the findings of our study. Karakurt et al. [20] measured Ang 1 and Ang 2 levels in the BM of 20 patients with ALL. We evaluated the expression of all members of the angiopoietin family (Ang1, Ang2, and Ang4) and receptors of this family (Tie1 and Tie2) in the BM of 40 ALL patients. Possible causes of inconsistencies in Ang1 expression levels may include differences in measurement methodology, number of samples, and leukemia type.
Another gene in the angiopoietin family is Ang2, which is a natural antagonist of Ang1 and increases vascular permeability and angiogenesis [10]. Our study showed a significantly increased expression of Ang2 in ALL patients compared to that observed in the control group. Numerous studies have been performed to evaluate the expression of this gene in hematological malignancies, all of which have shown an increase in patient Ang2 expression levels compared to the control group [20, 23, 30-32]. However, a study by Cheng et al. [28] reported decreased expression levels of Ang2 in MDS patients. Moreover, there was no statistically significant difference in the expression of Ang2 across demographic and clinical characteristics in the present study. In a study by Aref et al. [31], a correlation between increasing the number of blasts and increasing the serum amount of the Ang2 protein in AML patients was shown. Additionally, Schliemann et al. [32] have shown a direct correlation between Ang2 expression levels and the infiltration of leukemic blasts and an inverse association with the survival rate of AML patients. Ang2 also binds to a5b1 integrin, causing cell proliferation or migration, leading to the spread of leukemia [18].
The least studied member of the Ang-Tie system is Ang4. The precise function of Ang4 in angiogenesis has not yet been defined, due to the operational similarities of Ang4 with Ang1 [15]. However, the expression of Ang4 in the present study was not significantly different between the control and patient groups. Moreover, there was no statistically significant difference in its expression across patient demographics and clinical data.
In a study by Kivivuori et al. [33], no expression of the Tie1 gene in pediatric or adult ALL patients was reported. In contrast, our study showed a significant increase in Tie1 and Tie2 expression levels in newly diagnosed ALL patients. This discrepancy may be due to differences in methodology because we measured gene expression in the present study, whereas Kivivuori et al. investigated the presence of protein. Increased expression of the Tie1 receptor and its role in CLL development were first investigated by Aguayo et al. [34] in PB samples of patients with CLL using western blot analysis. Similar to our results, Verstovsek et al. demonstrated Tie1 upregulation in AML and CML patients [35, 36] and reported increased levels of Tie1 that were correlated with short survival of CML patients [36]. Increased levels of Tie1 may limit Ang1 function and consequently cause the spread of the tumor. Several studies have shown an increase in Tie2 gene expression in various hematological malignancies, which corroborates our findings [23, 28, 32]. Previous studies have also considered this increase in Tie2 gene expression as a marker of poor prognosis, especially in AML [16, 31, 37]. Notably, Schliemann et al. [32] reported that high expression of this receptor, along with the Ang2 gene, is directly related to leukemic blast infiltration and that it predicts short survival in AML patients. However, this increase in expression may lead to the production of soluble Tie2 (sTie2), which in turn may inhibit Ang1 activity [18]. However, there was no statistically significant association between Tie1 and Tie2 gene expression levels and patient demographics and clinical characteristics.
We performed ROC curve analysis to evaluate Ang-Tie system genes as diagnostic biomarkers in ALL samples. The combination of AUC values, sensitivity, and specificity of the Ang-Tie system genes can be considered as diagnostic biomarkers for patients with ALL. However, since we included only two types of ALL (pre-B and early pre-B-ALL), further studies with a large sample size that includes all types of ALL are needed to improve the sensitivity and specificity criteria of diagnostic markers. We recommend following up on the condition of patients after treatment to measure the effects of these proteins on prognosis, treatment, and survival. Furthermore, we believe it is crucial to investigate Ang4 expression in order to determine the role of this gene in hematological malignancies, including ALL. We also recommend the evaluation of the effects of angiopoietin and angiopoietin receptor suppressive drugs, such as pexmetinib or nilotinib.
In brief, we observed a significant decrease in Ang1 gene expression and an increase in Ang2, Tie1, and Tie2 gene expression. With increases in Ang2 and Tie2 levels, a decrease in Ang1 levels, and the possible generation of sTie2, ALL may progress towards exhibiting increased vascular permeability and angiogenesis. It is worth mentioning that since angiopoietins are secreted from both endothelial cells and malignant blasts, the blasts themselves may play a role in the spread of cancer.
ACKNOWLEDGMENTS
We thank all the individuals and their family members for their participation and support in this study.
REFERENCES
1. Siegel RL, Miller KD, Jemal A. 2020; Cancer statistics, 2020. CA Cancer J Clin. 70:7–30. DOI: 10.3322/caac.21590. PMID: 31912902.

2. Hunger SP, Mullighan CG. 2015; Acute lymphoblastic leukemia in children. N Engl J Med. 373:1541–52. DOI: 10.1056/NEJMra1400972. PMID: 26465987.

3. Terwilliger T, Abdul-Hay M. 2017; Acute lymphoblastic leukemia: a comprehensive review and 2017 update. Blood Cancer J. 7:e577. DOI: 10.1038/bcj.2017.53. PMID: 28665419. PMCID: PMC5520400.

4. Han Y, Wang X, Wang B, Jiang G. 2016; The progress of angiogenic factors in the development of leukemias. Intractable Rare Dis Res. 5:6–16. DOI: 10.5582/irdr.2015.01048. PMID: 26989643. PMCID: PMC4761589.

5. Aguayo A, Giles F, Albitar M. 2003; Vascularity, angiogenesis and angiogenic factors in leukemias and myelodysplastic syndromes. Leuk Lymphoma. 44:213–22. DOI: 10.1080/1042819021000029777. PMID: 12688336.

6. Allahbakhshian Farsani M, Kamel M, Mehrpouri M, et al. 2020; The expression of interferon gamma (IFN-g) and interleukin 6 (IL6) in patients with acute lymphoblastic leukemia (ALL). Pathol Oncol Res. 26:461–6. DOI: 10.1007/s12253-018-0536-z. PMID: 30443842.
7. Jabari M, Allahbakhshian Farsani M, Salari S, Hamidpour M, Amiri V, Mohammadi MH. 2019; Hypoxia-inducible factor1-A (HIF1a) and vascular endothelial growth factor-A (VEGF-A) expression in de novo AML patients. Asian Pac J Cancer Prev. 20:705–10. DOI: 10.31557/APJCP.2019.20.3.705. PMID: 30909668. PMCID: PMC6825791.
8. Eklund L, Kangas J, Saharinen P. 2017; Angiopoietin-Tie signalling in the cardiovascular and lymphatic systems. Clin Sci (Lond). 131:87–103. DOI: 10.1042/CS20160129. PMID: 27941161. PMCID: PMC5146956.

9. Davis S, Aldrich TH, Jones PF, et al. 1996; Isolation of angiopoietin-1, a ligand for the TIE2 receptor, by secretion-trap expression cloning. Cell. 87:1161–9. DOI: 10.1016/S0092-8674(00)81812-7. PMID: 8980223.

10. Maisonpierre PC, Suri C, Jones PF, et al. 1997; Angiopoietin-2, a natural antagonist for Tie2 that disrupts in vivo angiogenesis. Science. 277:55–60. DOI: 10.1126/science.277.5322.55. PMID: 9204896.

11. Harfouche R, Hasséssian HM, Guo Y, et al. 2002; Mechanisms which mediate the antiapoptotic effects of angiopoietin-1 on endothelial cells. Microvasc Res. 64:135–47. DOI: 10.1006/mvre.2002.2421. PMID: 12074640.

12. Khan AA, Sandhya VK, Singh P, et al. 2014; Signaling network map of endothelial TEK tyrosine kinase. J Signal Transduct. 2014:173026. DOI: 10.1155/2014/173026. PMID: 25371820. PMCID: PMC4211299.

13. Amani F, Allahbakhshian Farsani M, Gholami M, Aghamiri SMR, Bakhshandeh M, Hossein Mohammadi M. 2021; The protective effect of oleuropein against radiation-induced cytotoxicity, apoptosis, and genetic damage in cultured human lymphocytes. Int J Radiat Biol. 97:179–93. DOI: 10.1080/09553002.2020.1793014. PMID: 32970517.

14. Kim I, Kim JH, Moon SO, Kwak HJ, Kim NG, Koh GY. 2000; Angiopoietin-2 at high concentration can enhance endothelial cell survival through the phosphatidylinositol 3'-kinase/Akt signal transduction pathway. Oncogene. 19:4549–52. DOI: 10.1038/sj.onc.1203800. PMID: 11002428.

15. Lee HJ, Cho CH, Hwang SJ, et al. 2004; Biological characterization of angiopoietin‐3 and angiopoietin‐4. FASEB J. 18:1200–8. DOI: 10.1096/fj.03-1466com. PMID: 15284220.

16. Koenecke C, Kümpers P, Lukasz A, et al. 2010; Shedding of the endothelial receptor tyrosine kinase Tie2 correlates with leukemic blast burden and outcome after allogeneic hematopoietic stem cell transplantation for AML. Ann Hematol. 89:459–67. DOI: 10.1007/s00277-009-0869-5. PMID: 20054548.

17. Seegar TC, Eller B, Tzvetkova-Robev D, et al. 2010; Tie1-Tie2 interactions mediate functional differences between angiopoietin ligands. Mol Cell. 37:643–55. DOI: 10.1016/j.molcel.2010.02.007. PMID: 20227369. PMCID: PMC2841065.

18. Huang H, Bhat A, Woodnutt G, Lappe R. 2010; Targeting the ANGPT-TIE2 pathway in malignancy. Nat Rev Cancer. 10:575–85. DOI: 10.1038/nrc2894. PMID: 20651738.

19. Mazzieri R, Pucci F, Moi D, et al. 2011; Targeting the ANG2/TIE2 axis inhibits tumor growth and metastasis by impairing angiogenesis and disabling rebounds of proangiogenic myeloid cells. Cancer Cell. 19:512–26. DOI: 10.1016/j.ccr.2011.02.005. PMID: 21481792.

20. Karakurt N, Aksu T, Koksal Y, et al. 2016; Angiopoietins in the bone marrow microenvironment of acute lymphoblastic leukemia. Hematology. 21:325–31. DOI: 10.1080/10245332.2015.1125078. PMID: 26901808.

21. Hatfield KJ, Hovland R, Øyan AM, et al. 2008; Release of angiopoietin-1 by primary human acute myelogenous leukemia cells is associated with mutations of nucleophosmin, increased by bone marrow stromal cells and possibly antagonized by high systemic angiopoietin-2 levels. Leukemia. 22:287–93. DOI: 10.1038/sj.leu.2404985. PMID: 17943167.

22. Ghaffari S, Torabi-Rahvar M, Omidkhoda A, Ahmadbeigi N. 2019; Impact of various culture conditions on ex vivo expansion of polyclonal T cells for adoptive immunotherapy. APMIS. 127:737–45. DOI: 10.1111/apm.12981. PMID: 31273832.
23. Schliemann C, Bieker R, Padro T, et al. 2006; Expression of angiopoietins and their receptor Tie2 in the bone marrow of patients with acute myeloid leukemia. Haematologica. 91:1203–11. DOI: 10.1182/blood.V108.11.1894.1894. PMID: 16956819.

24. Müller A, Lange K, Gaiser T, et al. 2002; Expression of angiopoietin-1 and its receptor TEK in hematopoietic cells from patients with myeloid leukemia. Leuk Res. 26:163–8. DOI: 10.1016/S0145-2126(01)00110-2. PMID: 11755466.

25. Loges S, Heil G, Bruweleit M, et al. 2005; Analysis of concerted expression of angiogenic growth factors in acute myeloid leukemia: expression of angiopoietin-2 represents an independent prognostic factor for overall survival. J Clin Oncol. 23:1109–17. DOI: 10.1200/JCO.2005.05.058. PMID: 15718307.

26. Hou HA, Chou WC, Lin LI, et al. 2008; Expression of angiopoietins and vascular endothelial growth factors and their clinical significance in acute myeloid leukemia. Leuk Res. 32:904–12. DOI: 10.1016/j.leukres.2007.08.010. PMID: 17904634.

27. Handschuh L, Kaźmierczak M, Milewski MC, et al. 2018; Gene expression profiling of acute myeloid leukemia samples from adult patients with AML-M1 and-M2 through boutique microarrays, real-time PCR and droplet digital PCR. Int J Oncol. 52:656–78. DOI: 10.3892/ijo.2017.4233. PMID: 29286103. PMCID: PMC5807040.
28. Cheng CL, Hou HA, Jhuang JY, et al. 2011; High bone marrow angiopoietin-1 expression is an independent poor prognostic factor for survival in patients with myelodysplastic syndromes. Br J Cancer. 105:975–82. DOI: 10.1038/bjc.2011.340. PMID: 21878936. PMCID: PMC3185953.

29. Mahvi A, Mardani G, Ghasemi-Dehkordi P, et al. 2015; Effects of phenanthrene and pyrene on cytogenetic stability of human dermal fibroblasts using alkaline comet assay technique. Proc Natl Acad Sci India Sect B Biol Sci. 85:1055–63. DOI: 10.1007/s40011-015-0514-0.

30. Atesoglu EB, Tarkun P, Mehtap O, et al. 2016; Serum angiopoietin levels are different in acute and chronic myeloid neoplasms: angiopoietins do not only regulate tumor angiogenesis. Indian J Hematol Blood Transfus. 32:162–7. DOI: 10.1007/s12288-015-0548-8. PMID: 27065577. PMCID: PMC4789009.

31. Aref S, El Menshawy N, Azmy E, El-Refaie M. 2009; Soluble angiopoietin-2/sTie2 receptor ratio is an independent prognostic marker in adult acute myeloid leukemia. Indian J Hematol Blood Transfus. 25:17–22. DOI: 10.1007/s12288-009-0004-8. PMID: 23100966. PMCID: PMC3453480.

32. Schliemann C, Bieker R, Thoennissen N, et al. 2007; Circulating angiopoietin-2 is a strong prognostic factor in acute myeloid leukemia. Leukemia. 21:1901–6. DOI: 10.1038/sj.leu.2404820. PMID: 17597808.

33. Kivivuori SM, Siitonen S, Porkka K, Vettenranta K, Alitalo R, Saarinen-Pihkala U. 2007; Expression of vascular endothelial growth factor receptor 3 and Tie1 tyrosine kinase receptor on acute leukemia cells. Pediatr Blood Cancer. 48:387–92. DOI: 10.1002/pbc.20857. PMID: 16685739.

34. Aguayo A, Manshouri T, O'Brien S, et al. 2001; Clinical relevance of Flt1 and Tie1 angiogenesis receptors expression in B-cell chronic lymphocytic leukemia (CLL). Leuk Res. 25:279–85. DOI: 10.1016/S0145-2126(00)00139-9. PMID: 11248324.

35. Verstovsek S, Kantarjian H, Manshouri T, et al. 2002; Prognostic significance of Tie‐1 protein expression in patients with early chronic phase chronic myeloid leukemia. Cancer. 94:1517–21. DOI: 10.1002/cncr.10363. PMID: 11920509.

36. Verstovsek S, Estey E, Manshouri T, et al. 2001; High expression of the receptor tyrosine kinase Tie-1 in acute myeloid leukemia and myelodysplastic syndrome. Leuk Lymphoma. 42:511–6. DOI: 10.3109/10428190109064609. PMID: 11699417.

37. Lee CY, Tien HF, Hu CY, Chou WC, Lin LI. 2007; Marrow angiogenesis-associated factors as prognostic biomarkers in patients with acute myelogenous leukaemia. Br J Cancer. 97:877–82. DOI: 10.1038/sj.bjc.6603966. PMID: 17848952. PMCID: PMC2360422.

Fig. 1
Relative expression of Ang-Tie system genes (Ang1, Ang2, Ang4, Tie1, and Tie2) in patient samples and control samples.
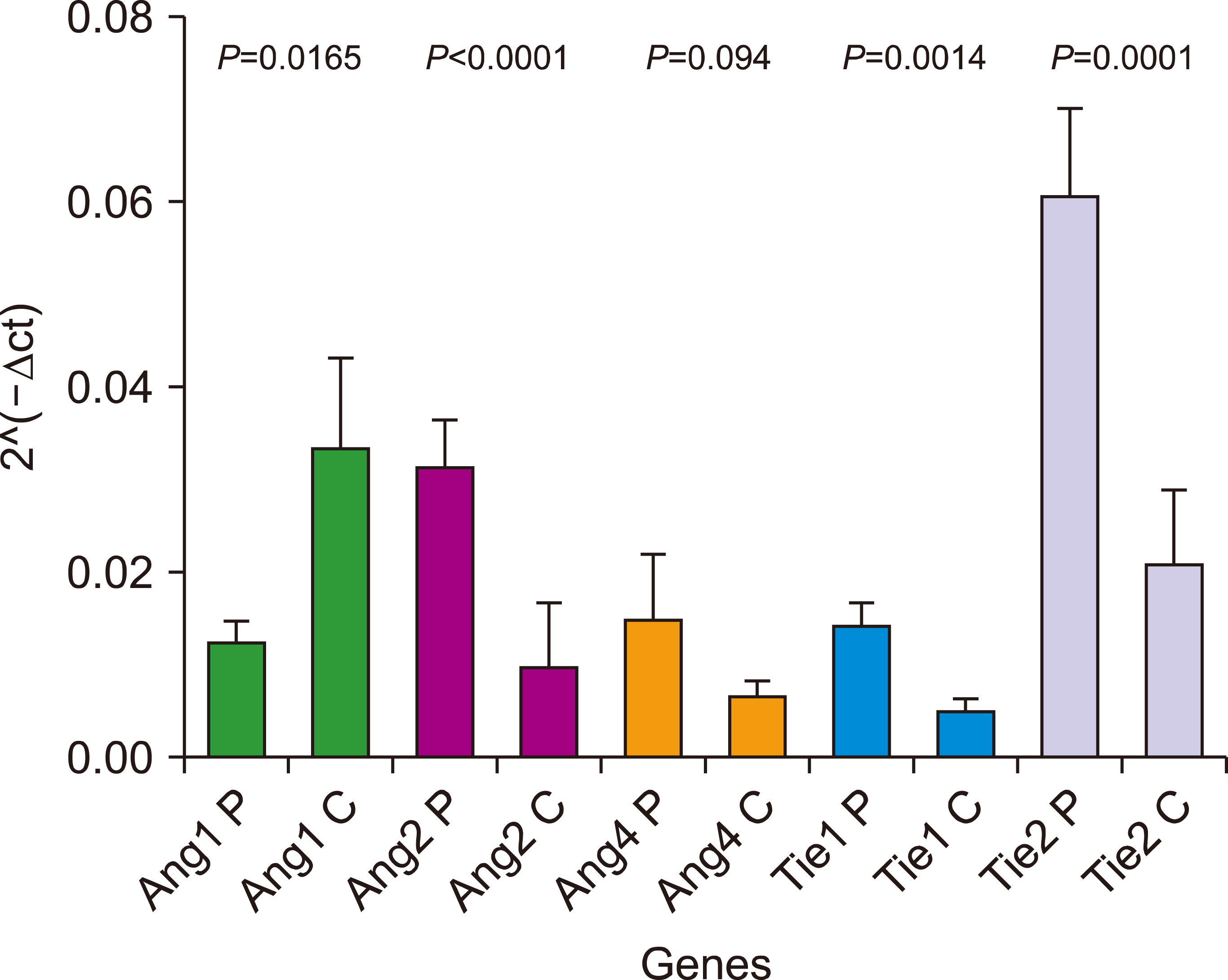
Fig. 2
Receiver operating characteristic curves of Ang1, Ang2, Tie1, and Tie2 genes for predicting their respective diagnostic potential in acute lymphoblastic leukemia.
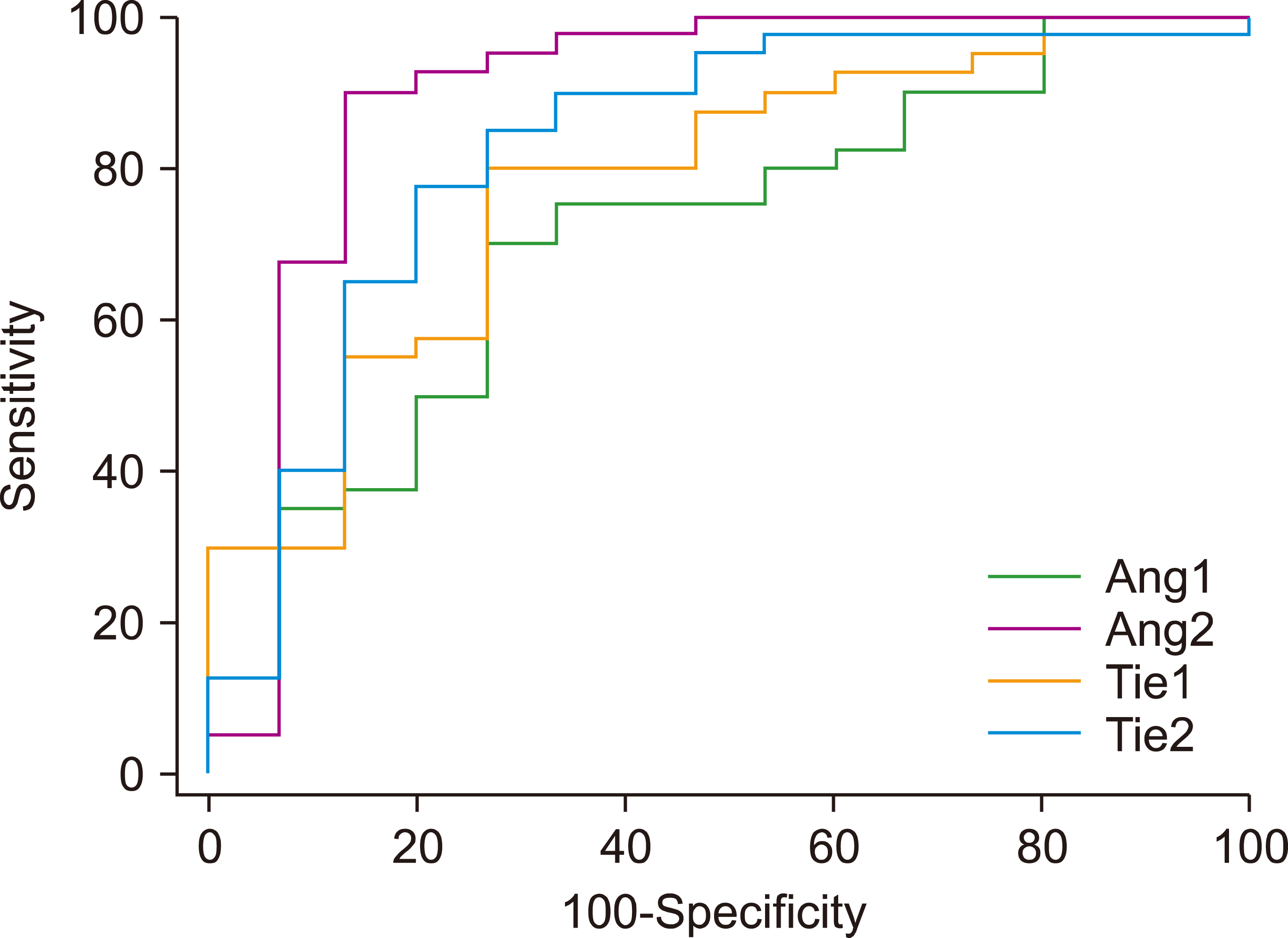
Table 1
The forward and reverse primer sequences and polymerase chain reaction product lengths used in this study.
Table 2
General baseline characteristics and clinical data of patients and controls.
Table 3
Ang1, Ang2, Agn4, Tie1, and Tie2 expression levels (means of ΔCt±standard deviation) according to the demographic and clinical data of the patients.
Table 4
Receiver operating characteristic curve analysis results.
| Genes | Estimate criterion | AUC | Ja) | Sensitivity | Specificity | P |
|---|---|---|---|---|---|---|
| Ang1 | ≤0.012 | 0.71 | 0.43 | 70.0 | 73.3 | 0.01 |
| Ang2 | >0.007 | 0.89 | 0.76 | 90.0 | 86.6 | <0.0001 |
| Tie1 | >0.005 | 0.77 | 0.53 | 80.0 | 73.3 | 0.0001 |
| Tie2 | >0.015 | 0.82 | 0.58 | 85.0 | 73.3 | <0.0001 |
| Genes combination | >0.045 | 0.71 | 0.38 | 85.0 | 53.3 | 0.01 |




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download