The current standard method for laboratory diagnosis of coronavirus disease (COVID-19) is molecular testing such as real-time reverse-transcription PCR (rRT-PCR). When molecular testing resources are insufficient, rapid antigen test (RAT) may be considered for massive and rapid testing [
1,
2]. The minimum performance requirements for a COVID-19 RAT compared with that of molecular testing have been suggested to be ≥90% clinical sensitivity and ≥97% specificity by the European Centre for Disease Prevention and Control (ECDC), and ≥80% clinical sensitivity and ≥97% specificity by the World Health Organization (WHO) [
1]. Nevertheless, the clinical performance of COVID-19 RATs is variable, which may hamper their clinical application [
3–
13].
Standard Q COVID-19 RAT (SQ-RAT; SD Biosensor, Suwon, Korea) was the first RAT that was approved by the Korean Ministry of Food and Drug Safety on November 11, 2020 (
https://mfds.go.kr/brd/m_74/view.do?seq=44004). We evaluated the performance of SQ-RAT and estimated its clinical sensitivity for COVID-19 diagnosis in the Korean population. This retrospective study was approved by the Institutional Review Boards (IRBs) of Jeonbuk National University Hospital (Jeonju), Seoul Medical Center (Seoul), and National Medical Center (Seoul), Korea (IRB approval numbers CUH-2020-12-022, SMC-2020-12-007, and NMC-2012-096, respectively). The requirement for patient consent was waived by all IRBs. All statistical analyses were performed using MedCalc Statistical Software v19.2.1 (MedCalc Software Ltd., Ostend, Belgium). SQ-RAT procedures were performed according to the manufacturer instructions, and collected data were interpreted and thoroughly checked by two laboratory physicians.
First, we determined the distribution of the initial threshold cycle (Ct) values for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection using rRT-PCR of upper respiratory tract specimens in the Korean population. The data were collected from the COVID-19 laboratory surveillance system of Korean Society for Laboratory Medicine from February 7, 2020 to December 17, 2020 [
14]. Data from four commercial rRT-PCR testing, including PowerChek 2019 nCoV (Kogene Biotech, Seoul, Korea), Allplex 2019-nCoV (Seegene, Seoul, Korea), Standard M nCoV (SD Biosensor), and Real-Q 2019-nCoV (Biosewoom, Seoul, Korea), all targeting the RNA-dependent RNA polymerase gene (
RdRp), were included in the analysis. The
RdRp Ct values were divided into six strata, and specimen sets were selected according to their distribution.
The
RdRp Ct distribution from 33,294 initial upper respiratory tract specimens collected at the time of COVID-19 diagnosis is shown in
Fig. 1A. Since 43.6% of the specimens had
RdRp Ct values lower than the limit of detection (LoD) of SQ-RAT, (manufacturer-claimed
RdRp Ct value of 23.37), this suggests that 43.6% of the patients had high viral loads and a diagnosis could be made by SQ-RAT, whereas the diagnosis could be missed for the majority (56.4%) of patients.
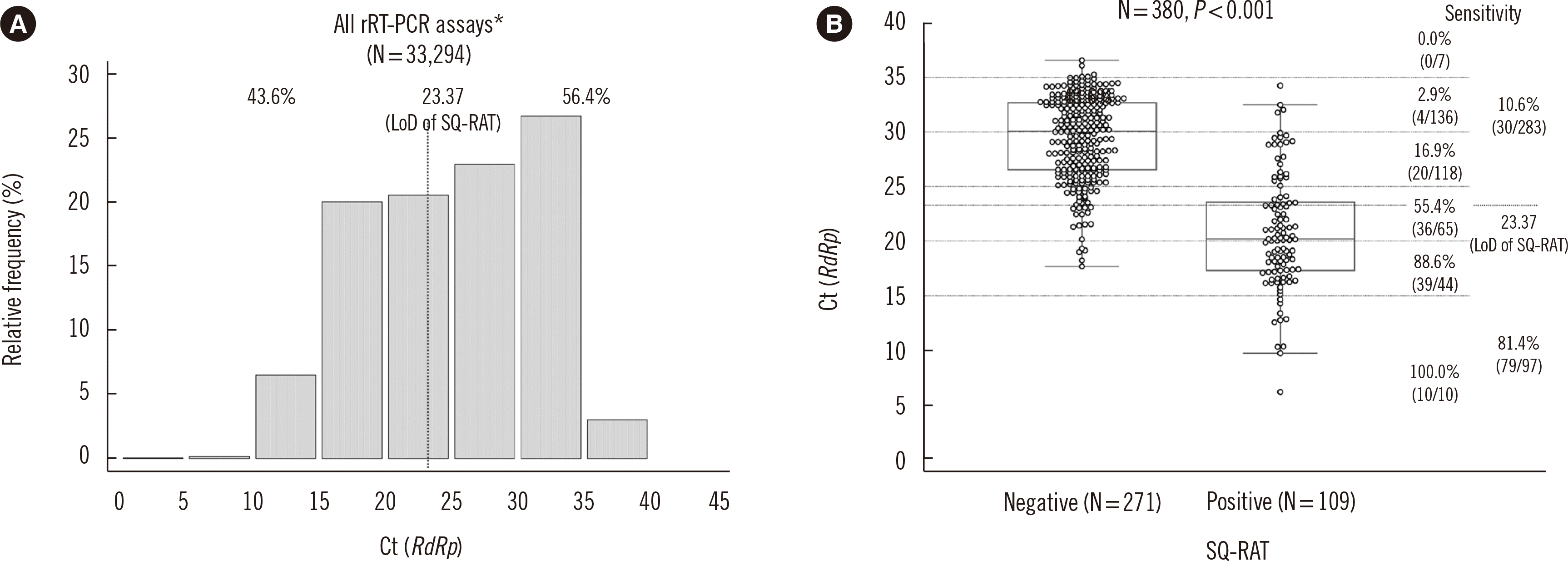 | Fig. 1
(A) Distribution of RdRp Ct values of all rRT-PCR tests performed on initial upper respiratory tract specimens obtained from newly diagnosed COVID-19 patients in Korea as of December 8, 2020. *Data were collected using the PowerChek 2019 nCoV (Kogene), Allplex 2019-nCoV (Seegene), Standard M nCoV (SD Biosensor), and Real-Q 2019-nCoV (Biosewoom) tests. (B) Clinical sensitivity of SQ-RAT compared with that of rRT-PCR by Ct stratum in specimens. The Ct data were collected using Standard M nCoV.
Abbreviations: COVID-19, coronavirus disease; Ct, threshold cycle; LoD, limit of detection; RdRp, RNA-dependent RNA polymerase gene; rRT-PCR, real-time reverse-transcription PCR; SQ-RAT, Standard Q COVID-19 rapid antigen test.

|
We tested residual nasopharyngeal swab specimens (collected from November 1 to November 30, 2020) with the Standard M nCoV Real-time Detection kit (SD Biosensor) according to the manufacturer’s instructions; those specimens were stored (–70°C) in universal transport medium (ASAN Transport Medium, Asanpharm, Seoul, Korea, or T-SWAB TRANSPORT Universal Transport Medium, Noble Biosciences, Hwaseong, Korea), after being used for SARS-CoV-2 rRT-PCR testing. We selected 380 SARS-CoV-2-positive specimens allocated to the six Ct strata and 300 SARS-CoV-2-negative specimens for SQ-RAT. The clinical sensitivity of SQ-RAT was 28.7% (109/380; 95% confidence interval [CI]: 24.2–33.5%), which decreased with increasing Ct value (
Fig. 1B). A significant difference in clinical sensitivity was observed between specimens with Ct values ≤23.37 (81.4%) and >23.37 (10.6%), which corresponds to the manufacturer-claimed LoD of SQ-RAT (
P<0.001, independent
t-test). The Ct values of SQ-RAT-positive and -negative specimens showed significant differences (
P<0.001, independent
t-test). The specificity of SQ-RAT was 100% (300/300; 95% CI: 98.8–100%).
As the conditions of residual specimens may affect RAT performance, the impact of freeze-thaw handling of upper respiratory tract specimens on the performance of SQ-RAT was evaluated using an independent set of 82 fresh upper respiratory tract specimens from COVID-19 patients. The specimens were tested using SQ-RAT within 24 hours of collection after confirming SARS-CoV-2 positivity by rRT-PCR. The specimens were frozen at –70°C for three days after initial testing, thawed at room temperature (20–25°C), and then retested using SQ-RAT within two hours. The SQ-RAT results before and after freezing showed 100% agreement for all 82 specimens, regardless of the initial Ct values. Thus, frozen specimens were acceptable for evaluating the performance of SQ-RAT (
Table 1).
Table 1
Comparison of SQ-RAT performance in fresh and frozen-thawed specimens
|
Ct (RdRp) |
SQ-RAT results |
|
|
Agreement |
Disagreement |
Sum |
|
|
Positive |
Negative |
|
5.0–14.9 |
11 |
0 |
0 |
11 |
|
15.0–19.9 |
13 |
0 |
0 |
13 |
|
20.0–24.9 |
6 |
7 |
0 |
13 |
|
25.0–29.9 |
0 |
6 |
0 |
6 |
|
30.0–34.9 |
0 |
10 |
0 |
10 |
|
Negative |
0 |
30 |
0 |
30 |
|
Sum |
30 |
53 |
|
83 |

Finally, we estimated the clinical performance of SQ-RAT based on the proportion of the observed Ct value distribution in the Korean population. The estimated clinical sensitivity of SQ-RAT in rRT-PCR-confirmed COVID-19 patients in Korea was 41.8% when the clinical sensitivity of each stratum was projected onto the Ct distribution data (
Fig. 1A). The estimated clinical sensitivity for each of the four rRT-PCR testing (
Table 2) varied from 33.8% to 59.7%.
Table 2
Estimated clinical sensitivity of SQ-RAT in comparison with four rRT-PCR tests
|
RdRp Ct stratum*
|
Sensitivity results |
Specimens of newly diagnosed COVID-19 patients with upper respiratory tract infection in Korea |
|
|
All rRT-PCR tests (%) (N = 33,294) |
Allplex (%) (N = 25,650) |
PowerChek (%) (N = 3,935) |
Standard M (%) (N = 1,942) |
Real-Q (%) (N = 1,762) |
|
Proportion of each stratum |
≤ 14.9 |
100 |
6.9 |
6.5 |
1.9 |
23.2 |
4.7 |
|
15.0–19.9 |
88.6 |
20.6 |
20.2 |
17.8 |
24.5 |
28.5 |
|
20.0–24.9 |
55.4 |
21.6 |
21.6 |
19.9 |
21.0 |
26.9 |
|
25.0–29.9 |
16.9 |
23.2 |
23.6 |
24.5 |
16.3 |
22.9 |
|
30.0–34.9 |
2.9 |
24.9 |
25.0 |
33.6 |
14.2 |
15.1 |
|
≥ 35.0 |
0 |
2.8 |
3.1 |
2.3 |
0.8 |
1.9 |
|
LoD of SQ-RAT |
≤ 23.37 |
81.4 |
42.0 |
41.1 |
33.4 |
62.9 |
52.7 |
|
> 23.37 |
10.6 |
58.0 |
58.9 |
66.6 |
37.1 |
47.3 |
|
Estimated clinical sensitivity of SQ-RAT*,†
|
|
41.8 |
41.1 |
33.8 |
59.7 |
49.2 |

We examined the clinical performance of SQ-RAT for COVID-19 diagnosis using the Ct values of initial upper respiratory tract specimens collected from newly diagnosed patients in Korea. SQ-RAT showed good specificity; however, its overall clinical sensitivity was low compared with that reported in previous studies using the same kit [
3,
4,
6,
8]. This discrepancy could be due to the difference in viral loads in the clinical specimens. Nevertheless, these findings were consistent with previous studies, in which the RAT showed good clinical sensitivity in specimens with high viral loads (Ct≤25), but not in specimens with low viral loads (Ct>25) [
16,
17]. Together, these findings suggest that the viral load distribution in the target population has a direct impact on the clinical sensitivity of RATs.
The Ct value distribution in this study was obtained from online laboratory surveillance data comprising Ct values collected at diagnosis and submitted by independent and hospital-associated clinical laboratories in Korea, but not from public health laboratories. More than a half of the specimens had Ct values>25 at diagnosis; consequently, the clinical sensitivity of SQ-RAT for COVID-19 diagnosis in the general Korean population was estimated to be low, which decreased in specimens with high Ct values (low viral loads). As Korea has implemented an aggressive testing strategy, the Korean surveillance data used in this study might include more pre-symptomatic or asymptomatic cases with low viral loads than data from other countries [
18,
19]. Several prospective evaluations of RATs in the general population reported a limited clinical performance. For example, the Liverpool pilot study reported that RATs would miss more than a half of the cases and that their clinical sensitivity did not fulfill the requirements of the ECDC and WHO [
1,
9,
10,
20]. These findings indicate that the viral loads of the target population should be considered for performance evaluation of
in vitro diagnostic testing for COVID-19, as the performance of a diagnostic method affects the effectiveness of measures to contain the COVID-19 pandemic [
13].
This study had some limitations. First, differences in the performance and usage of PCR reagents might have affected the Ct value distribution. Second, SQ-RAT was evaluated using residual upper respiratory tract specimens. Although most RAT evaluation studies used residual specimens, the clinical sensitivity may have been affected by the specimen type [
16,
17].
In summary, this study revealed that SQ-RAT has limited clinical sensitivity for COVID-19 diagnosis in the Korean population. Considering the sufficient molecular testing capacity in Korea, the usefulness of RATs for COVID-19 diagnosis seems to be limited to situations in which molecular testing cannot be accessed immediately.
Go to :






 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download