Abstract
Background
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is generally diagnosed by reverse transcription (RT)-PCR or serological assays. The SARS-CoV-2 viral load decreases a few days after symptom onset. Thus, the RT-PCR sensitivity peaks at three days after symptom onset (approximately 80%). We evaluated the performance of the ARCHITECT® SARS-CoV-2 IgG assay (henceforth termed IgG assay; Abbott Laboratories, Lake County, IL, USA), and the combination of RT-PCR and the IgG assay for COVID-19 diagnosis.
Methods
In this retrospective study, 206 samples from 70 COVID-19 cases at two hospitals in Tokyo that were positive using RT-PCR were used to analyze the diagnostic sensitivity. RT-PCR-negative (N=166), COVID-19-unrelated (N=418), and Japanese Red Cross Society (N=100) samples were used to evaluate specificity.
Results
Sensitivity increased daily after symptom onset and exceeded 84.4% after 10 days. Specificity ranged from 98.2% to 100% for samples from the three case groups. Seroconversion was confirmed from 9 to 20 days after symptom onset in 18 out of 32 COVID-19 cases with multiple samples and from another case with a positive result in the IgG assay for the first available sample.
Coronavirus disease 2019 (COVID-19) is a respiratory disease caused by the novel coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1]. The incubation period of SARS-CoV-2 ranges from one to 14 days, with the majority of symptoms manifesting in three to five days [2, 3].
COVID-19 is diagnosed by SARS-CoV-2 RNA detection in respiratory samples, especially in nasopharyngeal samples, using reverse transcription (RT)-PCR, and, serologically, by SARS-CoV-2 antibody detection in serum or plasma. The time from exposure is considered to have a strong influence on the sensitivity of RT-PCR assays; the viral load peaks three days after symptom onset, with a sensitivity of approximately 80% at this point [4–6]. Inconsistency of RT-PCR results was an issue for diagnosis in 12.5% and 21.4% of cases with COVID-19-like symptoms, showing a negative first and positive second RT-PCR results in two previous studies [7, 8]. Additionally, RT-PCR results performed intermittently often fluctuate between negative and positive [7].
In serological assays for SARS-CoV-2 antibodies, IgG and IgM are detected two to three weeks after symptom onset [5]. As of July 2020, 28 serological assay kits launched by 22 manufacturers have achieved Emergency Use Authorization by the U.S. Food and Drug Administration; among these, 21 are chemiluminescence microparticle immunoassays, including chemiluminescent immunoassays (CLIAs) and ELISAs [9].
The SARS-CoV-2 IgG assay and other serological assays have played important roles in the diagnosis and surveillance of COVID-19, but the clinical utility of such assays remains unclear. We hypothesized that COVID-19 diagnostic sensitivity may be improved by taking advantage of the difference in diagnostic sensitivity peaks between RT-PCR and serological assays with the combination of these methods.
We evaluated the performance of the ARCHITECT® SARS-CoV-2 IgG assay (henceforth termed IgG assay; Abbott Laboratories, Lake County, IL, USA) and the combination of RT-PCR and the IgG assay for COVID-19 diagnosis. We assessed results obtained by these methods in confirmed COVID-19 cases at two medical facilities in Tokyo retrospectively and longitudinally. This study can enrich the currently limited data on the IgG assay for Japanese samples.
This study was approved by the Ethics Committee of Faculty of Medicine, Toho University, Tokyo, Japan (No. A20028_A20020_A20014_A19099). Blood samples were collected from COVID-19 patients who agreed to participate in this study, on the day of admission, and approximately 3, 7, and 14 days after admission. Samples collected for routine laboratory tests were also analyzed. All serum and plasma samples for the serological assay were stored at –80°C before use. COVID-19 samples were stored for less than three months.
COVID-19 was diagnosed on the basis of SARS-CoV-2 RNA detection in a nasopharyngeal swab using RT-PCR. At Toho University, the laboratory-developed test (LDT) employing SARS-CoV-2 RT-PCR targets the N gene, which encodes the nucleocapsid protein [10]. Total RNA was extracted from nasopharyngeal swabs using QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) or BD MAXTM ExK TNA-3 (swab; Becton Dickinson, Franklin Lakes, NJ, USA). One-step RT-PCR was performed using QuantStudio® 5 (Thermo Fisher Scientific/Applied Biosystems, Waltham, MA, USA) or BD MAXTM Open System (BD) with TaqMan Fast Virus 1-Step Master Mix (Thermo Fisher Scientific) and BD MAXTNA MMK (SPC) one-step RT-PCR master mix. The primer set (NIID_2019-nCOV_N_F2 and NIID_2019-nCOV_N_R2) and a FAM-labeled probe (NIID_2019-nCOV_N_P2), which target the N gene, were used for amplification as previously reported [10]. The limits of detection (LODs) of the two LDT RT-PCR methods were evaluated using a 1×105/mL AccuPlex SARS-CoV-2 Verification Panel (SeraCare, Milford, MA, USA) progressively diluted with RPMI-1640 medium (Thermo Fisher Scientific) containing 1% (w/v) fetal bovine serum (Thermo Fisher Scientific). The LODs of the QuantStudio® 5 and BD MAX™ assays were 180 viral copies/mL (the verification kit was diluted 550 times) and approximately 330 viral copies/mL (the verification kit was diluted 300 times) in the initial sample, respectively. SARS-CoV-2 RT-PCR of samples from the National Hospital Organization Tokyo Medical Center was conducted as an administrative assay at a public research institution in Tokyo. RNA was extracted using QIAamp Viral RNA Mini Kit with a primer set and probe targeting the N and orf1ab genes as previously reported [11]. The LOD of the SARS-CoV-2 RT-PCR at the public research institution in Tokyo has not been published.
In total, 206 samples from 70 cases, including multiple samples available from 32 cases, were collected at Toho University Omori Medical Center and the National Hospital Organization Tokyo Medical Center, Tokyo, Japan, between March and May 2020. These samples were confirmed as positive for SARS-CoV-2 by RT-PCR.
We used three groups of samples for specificity evaluation. The first group comprised 166 samples from 109 cases of suspected COVID-19 judged to have a viral load below the LOD of RT-PCR for SARS-CoV-2. These samples were collected at Toho University Omori Medical Center and the National Hospital Organization Tokyo Medical Center between March and May 2020. The second group comprised 418 samples from 418 cases not suspected of having COVID-19 (collected at Toho University Omori Medical Center between March and June 2020). The third group comprised 100 samples collected in 2019 (prior to the COVID-19 outbreak) provided by the Japanese Red Cross (JRC).
Samples were analyzed using the IgG assay on an ARCHITECT® i2000SR analyzer (Abbott Laboratories) per the manufacturer’s instructions. The IgG assay is a fully automated CLIA for SARS-CoV-2 IgG detection in human serum or plasma. Samples, SARS-CoV-2 recombinant nucleocapsid antigen (N protein)-coated microparticles, and assay diluent were mixed for IgG binding. After washing, anti-human IgG acridinium-labeled conjugate was added to the mixture for binding to the SARS CoV-2 IgG captured on the microparticles. After washing, Pre-Trigger and Trigger Solutions (Abbott Laboratories) were added to start the chemiluminescent reaction, measured in relative light units and converted to the concentration of SARS-CoV-2 IgG in the sample. The cutoff index for positive and negative results was 1.40.
The sensitivity increased at ≤3, 4–6, 7–9, 10–12, 13–15, 16–18, and ≥19 days after symptom onset (Fig. 1). Of the three IgG assay-negative samples collected 16 days after symptom onset, two had index values close to the cutoff. These two samples were collected from the same patient (case 111) on days 18 and 19; a sample collected on day 20 was positive in the IgG assay (index value: 1.52; Fig. 1). The other sample (clearly negative) was collected from a patient with mild symptoms (case M42) 22 days after cough onset.
Assay specificities are shown in Fig. 2. In the group of non-COVID-19 (without an RT-PCR-positive history), the three IgG assay-positive samples were collected from the same patient (case 65). The IgG index in these samples were 3.36 on day 3, 3.47 on day 5, and 2.97 on day 8; the typical increase in the IgG index during the days after symptom onset was not observed, despite positivity in the IgG assay (Fig. 2).
In 18 of the 32 COVID-19 cases with longitudinal observations, seroconversion (i.e., serological assay result change from negative to positive) was observed from day 9 to 20, and 15 cases were tested positive within 12 days after symptom onset (Fig. 3A). The IgG index value peaked within at least 30 days after symptom onset in nearly all cases. The three cases with delayed seroconversion (case 11, 89, and 111) were not characterized by severity of symptoms (mild, no oxygen administration or ventilator required; moderate, oxygen administration required; and severe, ventilator required). In the 14 cases with the first sample being tested positive in the IgG assay, the IgG index values of five cases (20, 56, 91, 98, and 110) peaked on days 17–24 after symptom onset and then began to decline (Fig. 3B). None of these cases showed negative results in the IgG assay within approximately three months after symptom onset, and the largest decrease in the IgG index (from 9.62 to 7.09 in 43 days) was observed for case 98.
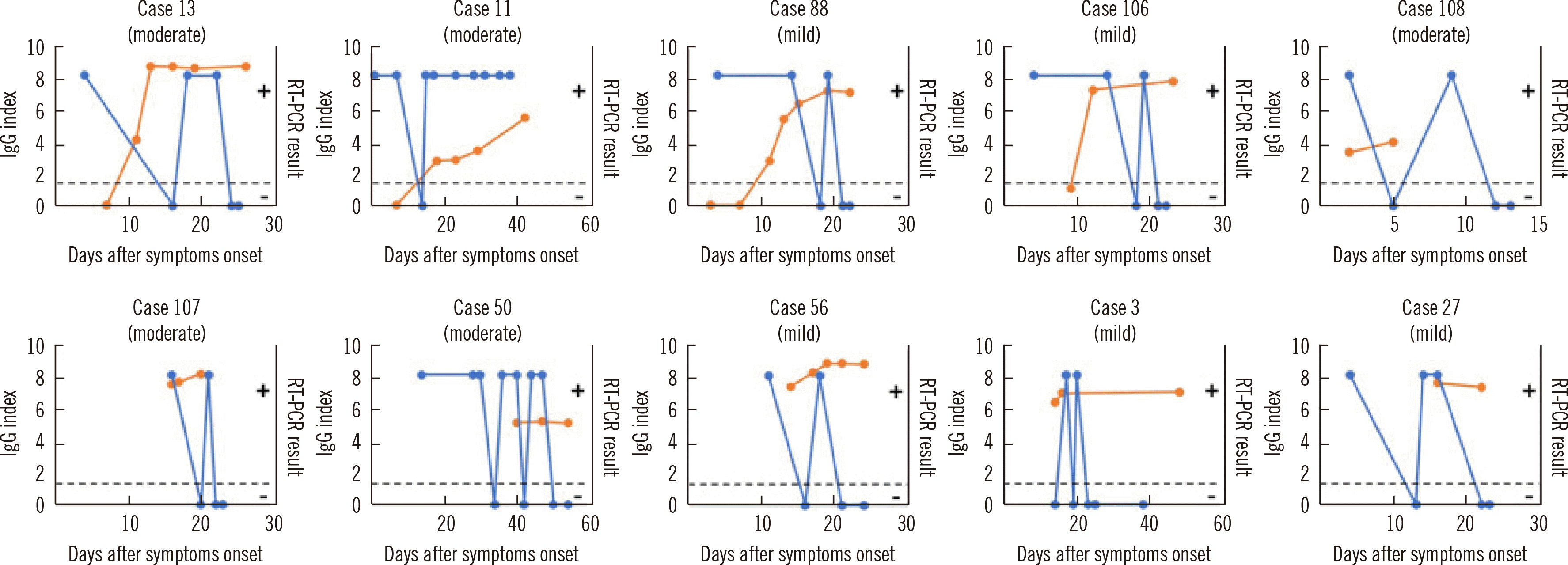
The RT-PCR and IgG assay results for available samples from 18 cases with seroconversion are shown in Fig. 4A. The duration from symptom onset to RT-PCR positivity ranged from 0 to 15 days (median: 5.5 days) and that from symptom onset to IgG assay positivity ranged from 9 to 52 days (median: 12 days). The interval between the first RT-PCR and IgG assay-positive results from symptom onset ranged from 2 to 51 days (median: 7 days). In 12 out of 18 cases, the RT-PCR result was positive on the same day or later than the day on which the IgG assay result was positive. In case 89, samples were not available for 51 days. Two cases (case 52 and 67) with the RT-PCR result changing from positive to negative within 10 days of symptom onset were positive for the IgG assay on day 10 after symptom onset (Fig. 4A). Nine days after symptom onset, varying RT-PCR results were observed for four cases (11, 13, 88, and 106), but the IgG assay result remained clearly positive (Fig. 4A). The longitudinal observations of RT-PCR and IgG assay results for 14 samples that tested positive in the IgG assay (collected 2 to 16 days after symptom onset) are shown in Fig. 4B. In 11 out of 14 cases, the RT-PCR results were positive on the same day or later than the day on which the IgG assay results were positive. Fluctuating RT-PCR results were observed in six cases (case 3, 50, 56, 91, 107, and 108; Fig. 4B). Of these 32 cases, RT-PCR and IgG assay results of four cases (case 67, 84, 86, and 97) were reversed (RT-PCR turned negative and the IgG assay turned positive) between 10 and 12 days after symptom onset. Fluctuating RT-PCR results (between negative and positive) were observed in 10 cases (Fig. 5).
We observed longitudinal results of SARS-CoV-2 RT-PCR and IgG assays after symptom onset retrospectively and determined the characteristics of each assay for COVID-19 diagnosis. The IgG assay showed more reliable longitudinal results than RT-PCR and compensated for the instability of RT-PCR for diagnosing COVID-19 using samples collected at least nine days after symptom onset.
Sensitivity analysis revealed more than 84.4% agreement between COVID-19 infection and SARS-CoV-2 IgG elevation 10 days after symptom onset; the SARS-CoV-2 IgG positivity increased daily (Fig. 1). Several SARS-CoV-2 serological studies in the USA using the ARCHITECT® platform have reported that 90% or more of the samples collected from COVID-19 cases needed to be collected more than 14 days after symptom onset to be positive in the IgG assay [12–14]. In this study, sensitivity was assessed by grouping the days after symptom onset into shorter periods than those in previous studies [12–14]. We found three IgG assay-negative samples even 16 days after symptom onset (samples indicated by “*” and “†” in Fig. 1). Case M42 involved mild COVID-19 symptoms 22 days after cough onset, and a nasopharyngeal swab performed at the same time tested positive by SARS-CoV-2 RT-PCR. Thus, we concluded that the initial cough was unrelated to SARS-CoV-2 infection and the patient had early-stage COVID-19 at the time of sampling. The apparently low IgG values in this sample also point to the possibility of a false-positive RT-PCR result.
The IgG assay specificity was very similar to the range of 99.35%–99.90% reported previously in the USA [12–14] (Fig. 2). The results for the three IgG assay-positive samples were probably false positives, because these samples were collected from the same patient (case 65), who tested negative in SARS-CoV-2 RT-PCR and the IgG index did not increase after symptom onset. Excluding this case, the assay specificity in the non-COVID-19 group was 100%. According to the IgG assay instruction manual (https://www.fda.gov/media/137910/download), four samples were suspected to have false-positive results among 1,070 samples assumed to be negative for COVID-19 (997 prior to the COVID-19 outbreak and 73 other respiratory diseases). Our data are similar to the manufacturer’s results.
As samples collected at least 10 days after symptom onset showed 84.4% sensitivity in the IgG assay, serological testing is useful for COVID-19 diagnosis. Assuming that the 32 patients with longitudinal data available for this study came to the hospital between 10 and 12 days after symptom onset, 19 cases could have been diagnosed by RT-PCR (those with positive RT-PCR results 10–12 days after symptom onset). A combination of RT-PCR and the IgG assay would facilitate a diagnosis of seven COVID-19 cases (case 52, 67, 68, 84, 86, 97, 108) in addition to those diagnosed using RT-PCR alone. Based on the SARS-CoV-2 viral load kinetics in previous reports [7, 8, 15] and in this study, RT-PCR results often fluctuate between positive and negative around the LOD. Therefore, the combination of RT-PCR and the IgG assay improved accuracy for COVID-19 diagnosis. RT-PCR compensates for the low sensitivity of the IgG assay, which in turn compensates for the fluctuation in RT-PCR results in the acute COVID-19 phase and the low sensitivity of RT-PCR in the late COVID-19 phase. Indeed, IgG assay positivity might be maintained for at least three months in a limited number of cases. Additionally, as an Italian report showed that 87.4% of patients recovering from COVID-19 had some persistent symptoms [16], the IgG assay might enable making a diagnosis for a patient with symptoms indicating COVID-19 persistence despite a negative RT-PCR result. .
Not all paired samples were collected at the same time or at predetermined intervals (e.g., hospitalization day 0, 3, 7, and 14). Therefore, we plan further evaluations using samples collected at the same time. We used only one IgG assay, which is a CLIA that uses the N protein to capture IgG in serum/plasma samples. Different results may be obtained using other SARS-CoV-2 IgG assays. Despite these limitations, this study provides important knowledge because only serological investigations using immunochromatography (also termed lateral-flow) methods have been reported for COVID-19 cases in Japan to date [17–19].
In conclusion, the combination of the SARS-CoV-2 RT-PCR and IgG assays improves the robustness for laboratory diagnosis during the course of COVID-19. In the near future, the IgG assay is expected to become one of the routine assays for COVID-19 diagnosis.
ACKNOWLEDGMENTS
We are grateful to Mr. Kenjiro Matsushita for help with the IgG assay. We also thank Mitchell Arico at Edanz Group (https://en-author-services.edanzgroup.com/ac) for editing a draft of this manuscript.
Notes
AUTHOR CONTRIBUTIONS
Aoki K, Takai K, and Tanaka I wrote the manuscript. Ishii Y conceptualized the study, supervised the work, was responsible for project administration, and reviewed the manuscript. Yoshimura T, Ishii T, Ishii Y, and Morita T supervised the study. Nagasawa T, Kodama N, and Shimodaira T performed assays. Kashiwagi K, Mori N, Matsubayashi K, Satake M, and Miyazaki T collected and provided samples. Tateda K was the principal investigator and acquired the research funding.
REFERENCES
1. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. 2020; A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 382:727–33. DOI: 10.1056/NEJMoa2001017. PMID: 31978945. PMCID: PMC7092803.

2. Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, et al. 2020; The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 172:577–82. DOI: 10.7326/M20-0504. PMID: 32150748. PMCID: PMC7081172.

3. Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. 2020; Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 382:1708–20. DOI: 10.1056/NEJMoa2002032. PMID: 32109013. PMCID: PMC7092819.

4. Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, et al. 2020; Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 323:1843–4. DOI: 10.1001/jama.2020.3786. PMID: 32159775. PMCID: PMC7066521.

5. Sethuraman N, Jeremiah SS, Ryo A. 2020; Interpreting diagnostic tests for SARS-CoV-2. JAMA. 323:2249–51. DOI: 10.1001/jama.2020.8259. PMID: 32374370.

6. Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J. 2020; Variation in false-negative rate of reverse transcriptase polymerase chain reaction-based SARS-CoV-2 tests by time since exposure. Ann Intern Med. 173:262–7. DOI: 10.7326/M20-1495. PMID: 32422057. PMCID: PMC7240870.

7. Li Y, Yao L, Li J, Chen L, Song Y, Cai Z, et al. 2020; Stability issues of RT‐PCR testing of SARS‐CoV‐2 for hospitalized patients clinically diagnosed with COVID‐19. J Med Virol. 92:903–8. DOI: 10.1002/jmv.25786.

8. Xiao AT, Tong YX, Zhang S. 2020; False negative of RT‐PCR and prolonged nucleic acid conversion in COVID‐19: rather than recurrence. J Med Virol. 92:1755–6. DOI: 10.1002/jmv.25855.

9. FDA, EUA Authorized Serology Test Performance. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/eua-authorized-serology-test-performance. Updated on 28th April 2021.
10. Shirato K, Nao N, Katano H, Takayama I, Saito S, Kato F, et al. 2020; Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV-2019) in Japan. Jpn J Infect Dis. 73:304–7. DOI: 10.7883/yoken.JJID.2020.061.
11. Nagashima M, Kumagai R, Yoshida I, Kawakami M, Nagano M, Asakura H, et al. 2020; Characteristics of SARS-CoV-2 isolated from asymptomatic carriers in Tokyo. Jpn J Infect Dis. 73:320–2. DOI: 10.7883/yoken.JJID.2020.137. PMID: 32350227.

12. Bryan A, Pepper G, Wener MH, Fink SL, Morishima C, Chaudhary A, et al. 2020; Performance characteristics of the Abbott Architect SARS-CoV-2 IgG assay and seroprevalence in Boise, Idaho. J Clin Microbiol. 58:727–8. DOI: 10.1128/JCM.00941-20. PMID: 32381641. PMCID: PMC7383515.

13. Theel ES, Harring J, Hilgart H, Granger D. 2020; Performance characteristics of four high-throughput immunoassays for detection of IgG antibodies against SARS-CoV-2. J Clin Microbiol. 58:e01243–20. DOI: 10.1128/JCM.01243-20. PMID: 32513859. PMCID: PMC7383546.

14. Tang MS, Hock KG, Logsdon NM, Hayes JE, Gronowski AM, Anderson NW, et al. 2020; Clinical performance of two SARS-CoV-2 serologic assays. Clin Chem. 66:1055–62. DOI: 10.1093/clinchem/hvaa120. PMID: 32402061. PMCID: PMC7239232.

15. Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. 2020; Virological assessment of hospitalized patients with COVID-2019. Nature. 581:465–9. DOI: 10.1038/s41586-020-2196-x. PMID: 32235945.

16. Carfì A, Bernabei R, Landi F. Gemelli Against COVID-19 Post-Acute Care Study Group. 2020; Persistent symptoms in patients after acute COVID-19. JAMA. 324:603–5. DOI: 10.1001/jama.2020.12603. PMID: 32644129. PMCID: PMC7349096.

17. Imai K, Tabata S, Ikeda M, Noguchi S, Kitagawa Y, Matuoka M, et al. 2020; Clinical evaluation of an immunochromatographic IgM/IgG antibody assay and chest computed tomography for the diagnosis of COVID-19. J Clin Virol. 128:104393. DOI: 10.1016/j.jcv.2020.104393. PMID: 32387968. PMCID: PMC7191278.

18. Takita M, Matsumura T, Yamamoto K, Yamashita E, Hosoda K, Hamaki T, et al. 2020; Geographical profiles of COVID-19 outbreak in Tokyo: an analysis of the primary care clinic-based point-of-care antibody testing. J Prim Care Community Health. 11:2150132720942695. DOI: 10.1177/2150132720942695. PMID: 32674696. PMCID: PMC7370565.

19. Lisboa Bastos M, Tavaziva G, Abidi SK, Campbell JR, Haraoui LP, Johnston JC, et al. 2020; Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 370:m2516. DOI: 10.1136/bmj.m2516. PMID: 32611558. PMCID: PMC7327913.

Fig. 1
Sensitivity of the ARCHITECT® SARS-CoV-2 IgG assay for COVID-19 diagnosis. The SARS-CoV-2 IgG index is plotted according to the number of days after symptom onset for 206 samples collected from 70 confirmed COVID-19 cases. *Samples collected from the same patient (case 111) on days 18 and 19 and a sample collected on day 20 were positive in the IgG assay (index value: 1.52). †Sample collected from a patient (case M42) with mild symptoms at 22 days after cough onset and a nasopharyngeal swab obtained at the same time tested positive by the SARS-CoV-2 RT-PCR assay. The dashed line indicates the IgG assay cutoff value (index=1.40).
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; COVID-19, Coronavirus disease 2019; RT-PCR, reverse transcription-PCR.
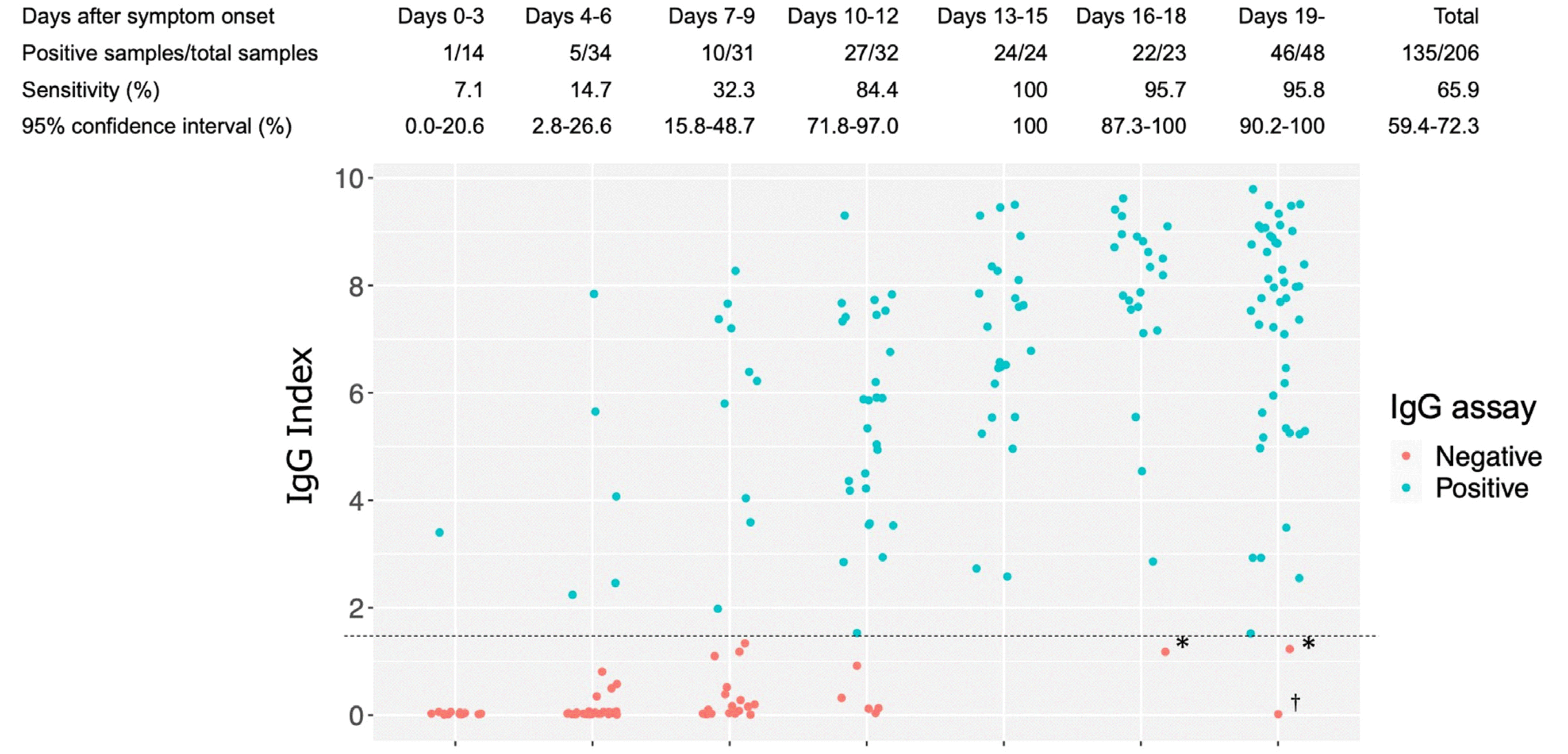
Fig. 2
Specificity of the ARCHITECT® SARS-CoV-2 IgG assay for COVID-19 diagnosis. IgG assay plot of cases and controls, including non-COVID-19 cases and cases not suspected to be COVID-19. “Non-COVID-19”: non-COVID-19 samples collected at Toho University Omori Medical Center and the National Hospital Organization Tokyo Medical Center between March and May 2020; “COVID-19 unsuspected”: samples not suspected of COVID-19 collected at Toho University Omori Medical Center; “JRC pre-COVID-19 outbreak”: blood donation samples provided by the Japanese Red Cross Society. Three IgG assay-positive samples collected at Toho University Omori Medical Center, which were non-COVID-19, were collected from one patient (case 65). The dashed line indicates the IgG assay cutoff value (index=1.40).
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; COVID-19, Coronavirus disease 2019; JRC, Japanese Red Cross Society.
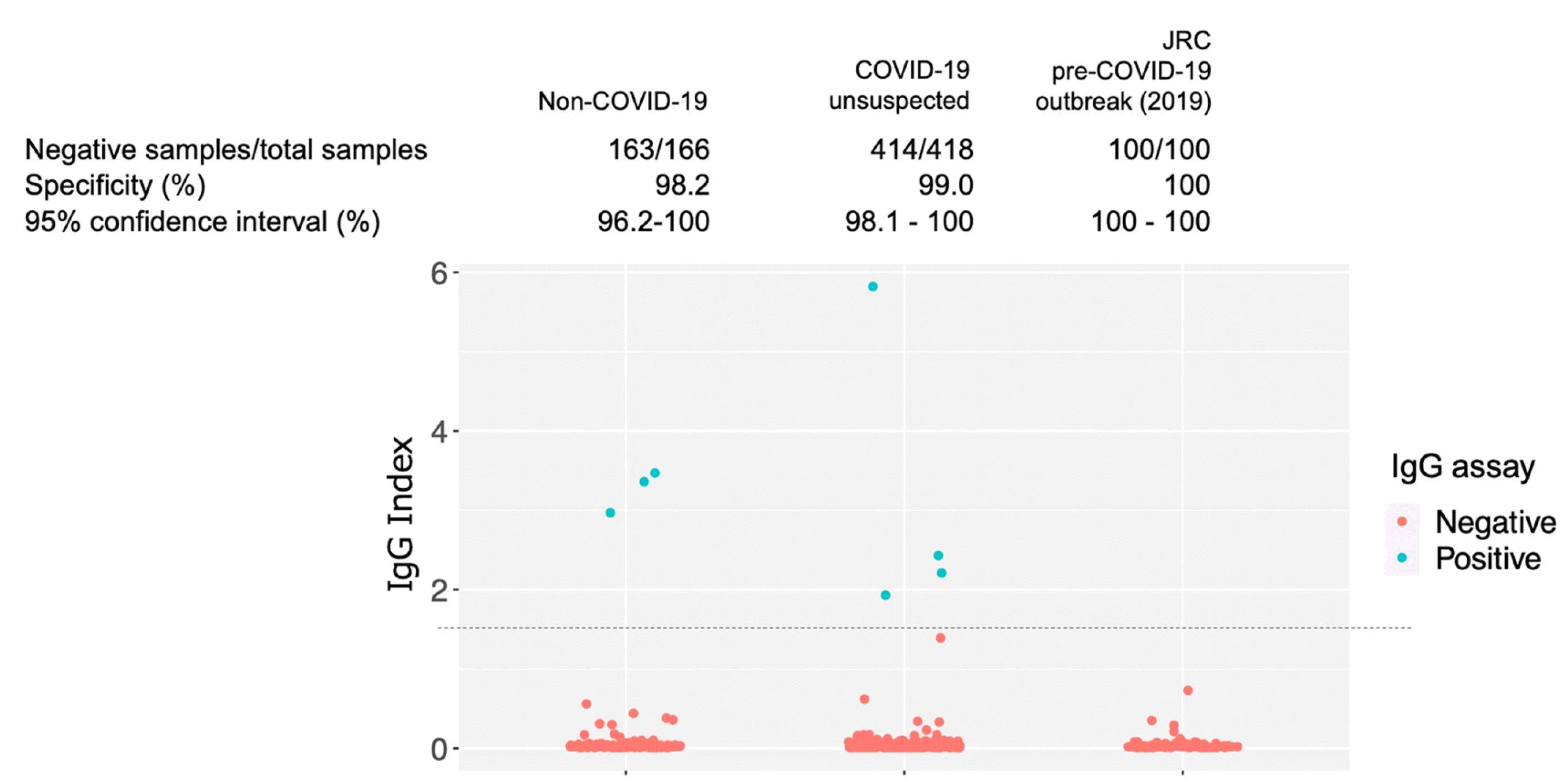
Fig. 3
Longitudinal observation of SARS-CoV-2 IgG assays in COVID-19 cases. (A) Changes in the SARS-CoV-2 IgG index in 18 COVID-19 cases that showed seroconversion (i.e., serological assay result change from negative to positive). (B) Longitudinal observation of SARS-CoV-2 IgG in 14 COVID-19 cases positive for SARS-CoV-2 IgG in the first antibody assay. The dashed line indicates the IgG assay cutoff value (index=1.40).
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; COVID-19, Coronavirus disease 2019.
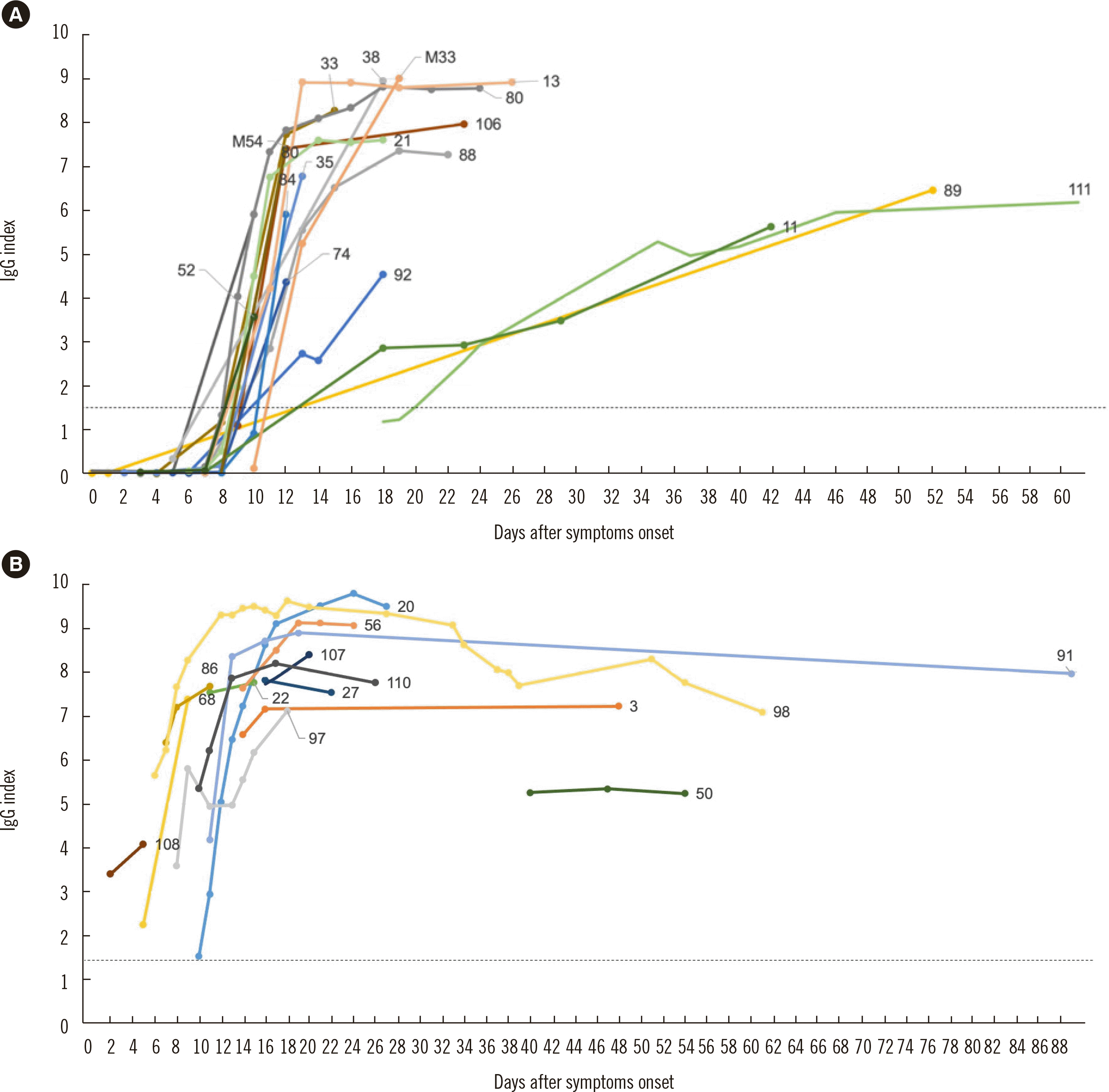
Fig. 4
Longitudinal observation of SARS-CoV-2 RT-PCR and IgG assays in COVID-19 cases. (A) RT-PCR results and the IgG assay index value for the cases shown in Fig. 3A (seroconversion observed). (B) RT-PCR results and the IgG assay index values for the cases shown in Fig. 3B (positive for IgG in the first antibody assay).
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; COVID-19, Coronavirus disease 2019; RT-PCR, reverse transcription-polymerase chain reaction.
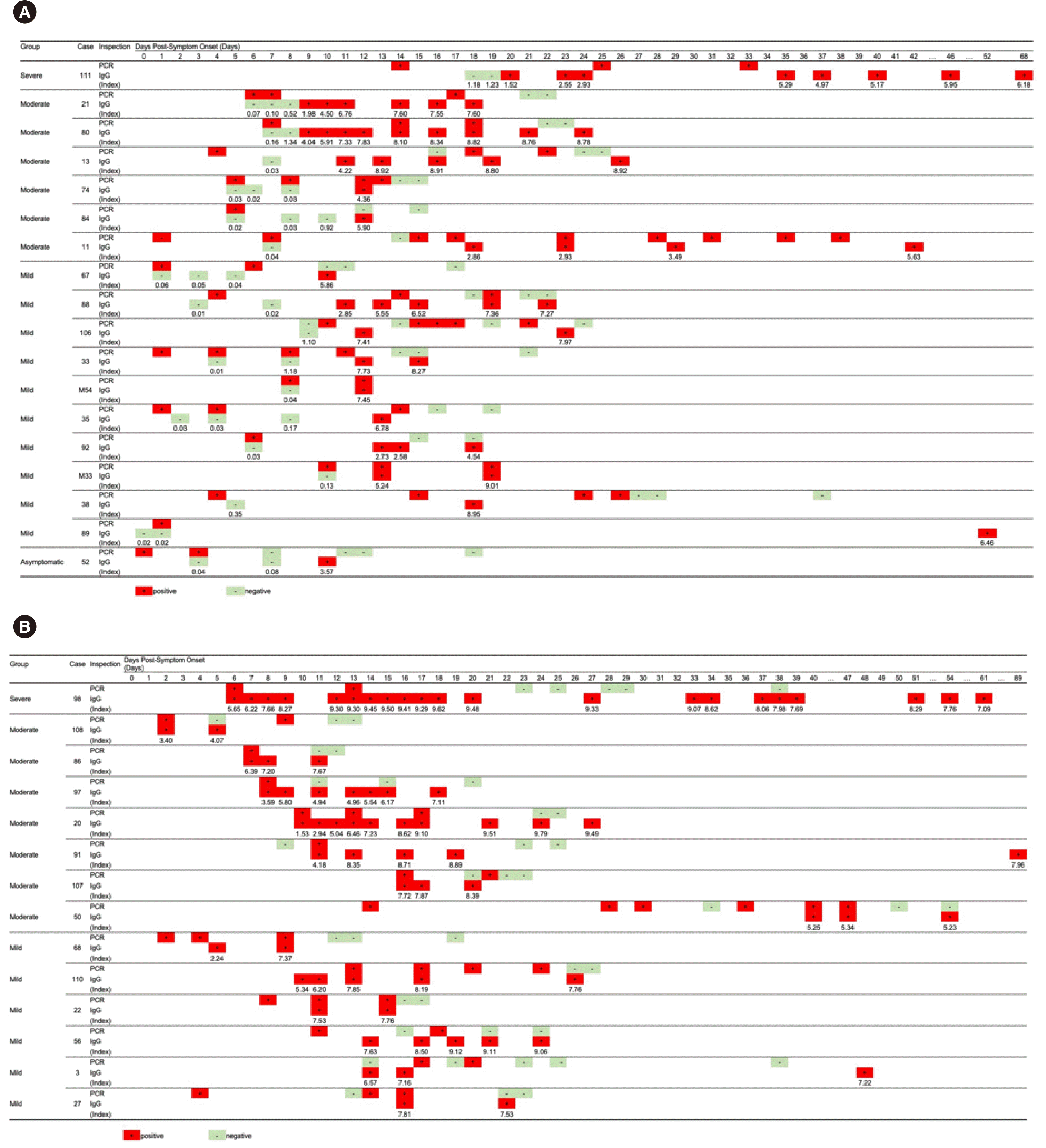
Fig. 5
Longitudinal observation of SARS-CoV-2 RT-PCR and IgG assays in COVID-19 cases with fluctuating RT-PCR results. Orange dots indicate IgG results and the dashed line shows the IgG assay cutoff value (index=1.40). Blue dots indicate RT-PCR results (positive and negative).
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; COVID-19, Coronavirus disease 2019; RT-PCR, reverse transcription-polymerase chain reaction.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download