Introduction
Why Is the S Protein the Target of Most COVID-19 Vaccine Designs?
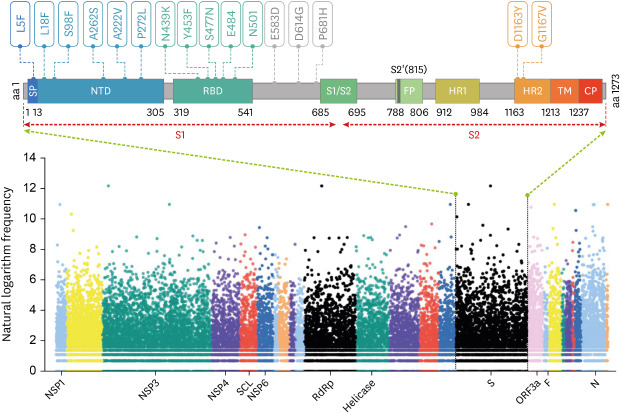
Fig. 1
The distribution of genome-wide mutations on SARS-CoV-2 and the key mutations in S protein. The data of mutations in hCoV-19 genomes was obtained from GISAID database from January 1, 2020, to December 26, 2020. The Y-axis represents the natural logarithm frequency of each mutation in the whole genome of SARS-CoV-2. The X-axis represents the name of the marker gene or protein, and the relative positions of other unlabeled genes or proteins can be found in the GISAID database. Each mutation was shown as a solid dot colored by Open Reading Frame of hCoV-19 genome. The S protein structure was presented following a previous study9 (Wrapp et al. Science 2020;367:1260-3). The Arabic numerals below the S protein structure represent the amino acid site in the full-length amino acid sequence.
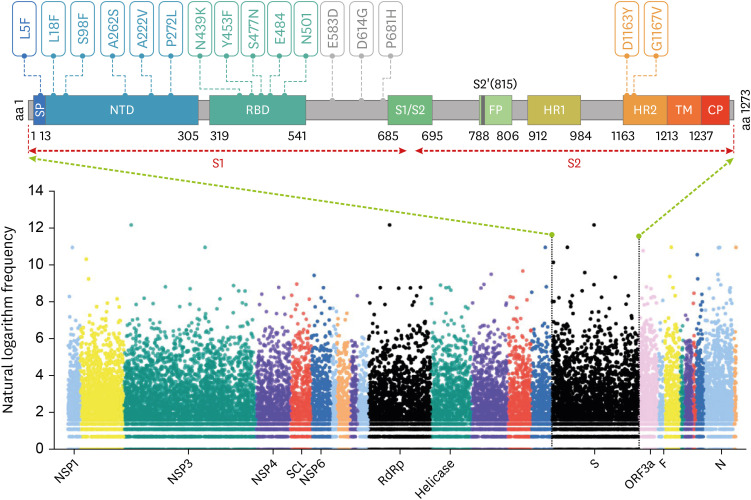
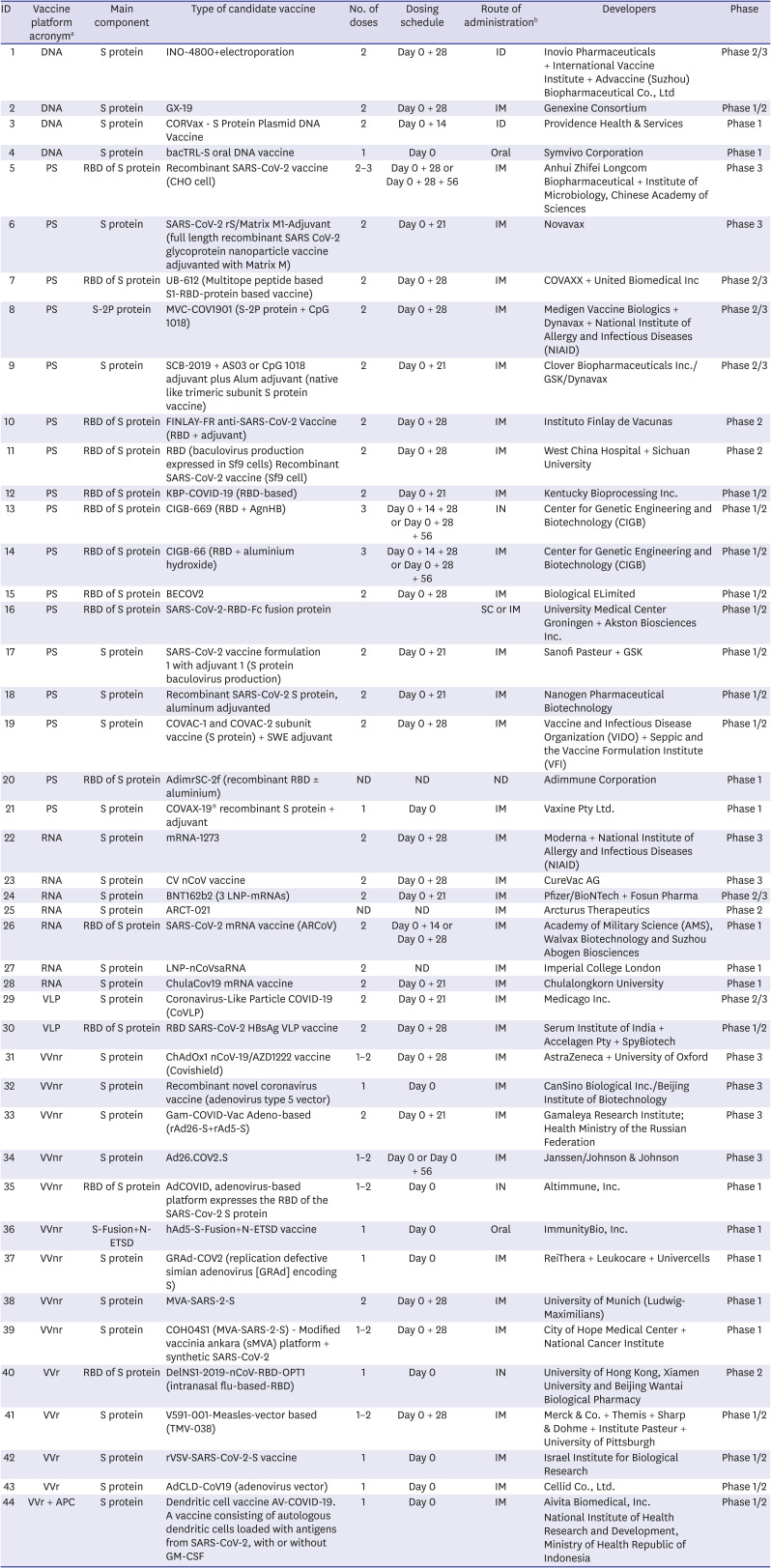
Table 1
Landscape of clinical development of COVID-19 candidate vaccines based on S protein
What Are the Key Mutations of the S Protein?
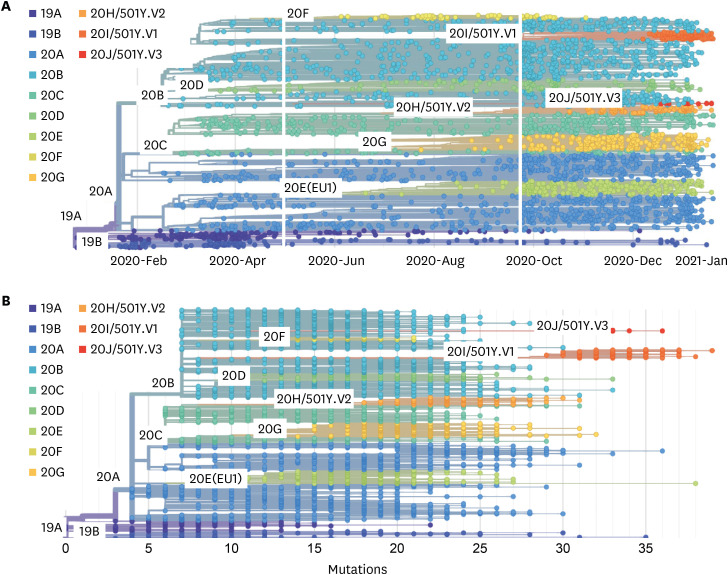
Fig. 2
The phylogeny and mutations of SARS-CoV-2 viruses. This phylogeny shows evolutionary relationships of SARS-CoV-2 viruses from the ongoing COVID-19 pandemic (A). Although the genetic relationships among sampled viruses are quite clear, there is considerable uncertainty surrounding estimates of specific transmission dates and in the reconstruction of geographic spread. The number of mutations in hCoV-19 genomes was obtained from Nextstrain database and showed by solid dot (B). Compiled Nextstrain SARS-CoV-2 resources are available at https://nextstrain.org/sars-cov-2/. All of data was obtained on January 26, 2021.
Do These Mutations Affect COVID-19 Vaccines Targeting the S Protein?
What Measures Should Be Taken to Deal with Viral Mutations?
(1) The monoclonal antibodies employed in the clinic should be tested again against the new variants. As a COVID-19 therapeutic agent, S protein-specific neutralizing antibodies can interact with the SARS-CoV-2 S protein to inhibit the virus from invading the human body. Monoclonal antibodies have been authorized for emergency use. It has been reported that viral mutants may reduce the effectiveness of neutralizing antibodies.29 Therefore, it is urgent to confirm the efficacy of these monoclonal and serum antibodies in neutralizing mutant viruses.
(2) Vaccines under EUA may need to be updated periodically with respect to clinical efficacy against SARS-CoV-2 variants. Currently, 44 vaccines that have entered the clinical trial stage were developed based on the S protein, and the data indicate that the S protein is the most mutated part of the SARS-CoV-2 virus. Increasing evidence also shows that some COVID-19 vaccines are less effective in protecting against variants.2428 These data suggest that vaccine manufacturers must update their vaccines in time to deal with viral mutations. Otherwise, the efficacy of the COVID-19 vaccine may be affected.
(3) The effectiveness of vaccines under EUA should be evaluated against SARS-CoV-2 variants. Phase III clinical trials of the six vaccines currently approved for emergency use were completed before the end of 2020. The actual efficacy data were also obtained based on infection by the original SARS-CoV-2 strain. This means that the efficacy of these vaccines against SARS-CoV-2 variants remains unknown. Thus far, only the efficacy of the NVX-CoV2373 vaccine against variants has been studied in a phase III clinical trial, and the results indicate that the efficacies of the NVX-CoV2373 vaccine against the original COVID-19, the UK variant, and the South African variant strains were 95.6%, 85.6%, and 49.4%, respectively.28
(4) For vaccines still in pre-clinical studies, SARS-CoV-2 variant-infected animal models should be established to assess their efficacy. Transgenic mice with humanized lungs and immune systems represent ideal animal models for COVID-19 vaccine and drug development.30 In August 2020, Hansen et al.31 created a humanized mouse model and compared the similarities and consistencies of antibodies against the SARS-CoV-2 S protein produced by humanized mice and convalescent patients. In the future, the development of a transgenic or humanized animal model for SARS-CoV-2 variant infection will greatly promote the development of more effective COVID-19 vaccines.
(5) Implement more stringent public health control strategies, such as wearing masks and social distancing. The scientific formulation and rapid and effective implementation of public health control strategies by governments are decisive factors for decreasing viral transmission, especially in countries where vaccines have not yet been made available. If people strictly abide by these public health control strategies, SARS-CoV-2 transmission would decrease, thereby lowering the frequency of mutations. In the face of the rapidly mutating virus, formulating and following public health control strategies is one of the most effective and economical strategies to deal with such crises.
(6) Efforts should be made to curb the spread of variant SARS-CoV-2 strains through international cooperation and by strengthening immigration quarantine. Experience and studies indicate that the rapid spread of COVID-19 is closely related to intensive personnel exchanges and cross-border travel.32 What is even more worrying is that variant SARS-CoV-2 viruses tend to have stronger transmission capabilities, which makes the spread of such variants more difficult to control internationally. To circumvent this challenge, effective international cooperation strategies should be formulated and implemented, and at the same time, countries should strengthen their immigration quarantine policies to reduce the risk of the cross-border transmission of SARS-CoV-2 variants.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download