INTRODUCTION
Gallstone disease is prevalent in many regions of the world and can occur in any location in the human biliary tract, including gallbladder, extrahepatic duct, or intrahepatic duct.
1 Choledocholithiasis is defined as the presence of stone inside the common bile duct (CBD), is commonly encountered, and the leading cause of hospitalization due to a gastrointestinal disorder.
2 At least 15% of patients with cholelithiasis have concurrent choledocholithiasis.
34 In the West, the majority of CBD stones are composed of cholesterol stones formed in the gallbladder. In fact, fewer than 10% of CBD stones are formed de novo within the CBD.
4 However, in the East, the incidence of primary choledocholithiasis (the formation of stones within the CBD) is much higher than in the West because of greater incidences of chronic biliary tree infection and infestation, and these stones are almost all pigment stones and frequently cause bile duct inflammation, that is, recurrent pyogenic cholangitis. These stones are composed of calcium bilirubinate and are associated with biliary cirrhosis and cholangiocarcinoma.
5 The precise pathophysiology of pigment stone is unknown, but infections of bile ducts by parasites like liver flukes and parasitic worms and biliary bacteria are considered by some to be responsible.
6 Bile stasis induced by parasites and their ova has been postulated to lead to bacterial colonization and hydrolysis of bilirubin conjugates and phospholipids by bacterial enzymes and that the insoluble free bilirubin and fatty acid formed precipitate with biliary calcium. The resulting concretions are termed pigment stones.
6 Many bacteria, such as
Escherichia coli,
Klebsiella pneumoniae,
Enterococcus faecium,
Enterobacter cloacae, and
Pseudomonas aeruginosa, have been identified in bile or gallstone samples by cultivation or polymerase chain reaction (PCR) and proposed to be associated with stone formation.
78 However, culture-based methods have been demonstrated to be insensitive and biased in terms of identifying bacteria related to CBD stone development.
8
Recent advances in technology, such as cultivation and bacteria-specific PCR and next-generation sequencing (NGS) have been used to identify and analyze components of microbiomes and have substantially improved our understanding of microbial communities associated with human disease.
9 In particular, NGS has proven to have advantages for microbiome studies in multiple human body sites, such as studies of the gastrointestinal tract, oral cavity, nasal cavity, skin, and vagina.
101112 However, a few studies conducted on fecal samples from gallstone patients showed they exhibited microbiota dysbiosis.
1314 No suitable NGS study has been conducted on the microbiome in the biliary tree or on the effect of the microbiome on gallstone occurrence.
14 Therefore, we conducted an NGS-based study using 16S sequencing to analyze the bile samples of 28 patients with obstructive biliary disease to gain a clearer picture of the structures and components of microbial communities of the biliary tract in these patients and to determine the characteristics of the biliary tract core microbiome and its potential relations with the pathogenesis of choledocholithiasis.
METHODS
Patients and sample collection
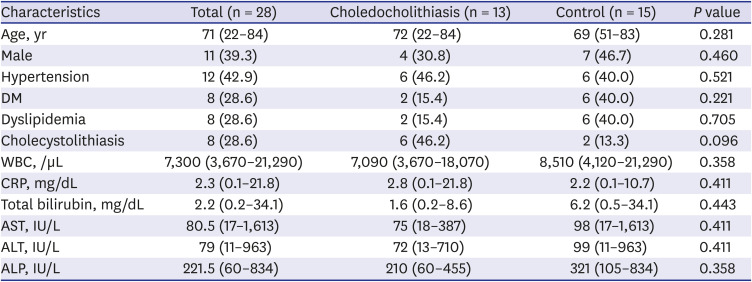
From February 2019 to January 2020, patients with a diagnosis of biliary disease, including cholangiocarcinoma, new-onset CBD stone, and benign biliary stricture were recruited at our institution. The study inclusion criteria were as follows: 1) diagnosis of biliary disease with evidence by abdominal imaging (abdominal ultrasound, computed tomography, or magnetic resonance imaging); 2) the need for endoscopic retrograde cholangiopancreatography (ERCP) for bile duct decompression; and 3) a naïve ampulla. The exclusion criteria applied were: 1) inability or refusal to give written informed consent, 2) indication for pancreatic ERCP, 3) previous procedure performed at the ampulla, 4) confirmed cholesterol stone or recurrent CBD Stone after ERCP, and 5) a problematic endoscopic approach (e.g., due to esophageal stenosis, gastric outlet obstruction, or duodenal stenosis). Prior to ERCP, all patients were administrated prophylactic antibiotics (ciprofloxacin 400 mg IV over 60 minutes) for prevention of bacteremia. ERCP was performed using a conventional side-viewing duodenoscope (TJF-260; Olympus Corporation, Tokyo, Japan) and a straight standard injection catheter. After achieving the therapeutic aim of ERCP, an endoscopic nasobiliary drainage (ENBD) tube was inserted and its proximal end was lodged at the proximal CBD. Bile samples (20–30 cc) were aspirated 24 hours after endoscopic procedures from CBDs via ENBDs to prevent contamination of the upper gastrointestinal tract including the oral cavity. Samples were immediately placed in germ-free sputum cups and stored at −80°C until required.
DNA extraction from bile samples
The total bacterial genomic DNA extraction from 10 mL bile acid samples was performed using a Maxwell® RSC PureFood GMO and Authentication Kit (Promega, Madison, WI, USA). Samples were initially centrifuged at 5,000 g at room temperature for 5 minutes and the pellets obtained were resuspended in 500 µL of cetyltrimethylammonium bromide buffer, according to the manufacturer's instructions. DNA concentrations were calculated using a UV-vis spectrophotometer (NanoDrop 2000c; Thermo Fisher Scientific, Waltham, MA, USA) and quantified using a QuantiFluor® ONE dsDNA System (Promega). DNA samples were stored at −20°C until required for experiments.
PCR amplification of the V3–V4 region of the bacterial 16S ribosomal RNA (rRNA) gene
The V3–V4 region of the bacterial 16S rRNA gene was amplified from extracted DNA using the F319 (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACG-GGNGGCWGCAG) and R806 (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACA-GGACTACHVGGGTATC-TAATCC-3′) primer sets. PCR products were subjected to 2% agarose gel electrophoresis and purified using AMPure XP magnetic beads (Beckman Coulter, Wycombe, UK). The qualities of purified amplicons were determined using a Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). Secondary amplification was then performed over 8 cycles to attach Illumina Nextera barcodes (Illumina, Inc., San Diego, CA, USA) using the i5 forward primer and the i7 reverse primer. Amplified products were purified using AMpure XP magnetic beads (Beckman Coulter) according to the manufacturer's protocol. Purified amplicons were quantified using a QuantiFluor® ONE dsDNA System (Promega). Amplicon size and quality were evaluated using a Bioanalyzer 2100. Pooled libraries were sequenced using an Illumina MiSeq instrument using a MiSeq v3 Reagent Kit (Illumina, Inc.).
Data analysis
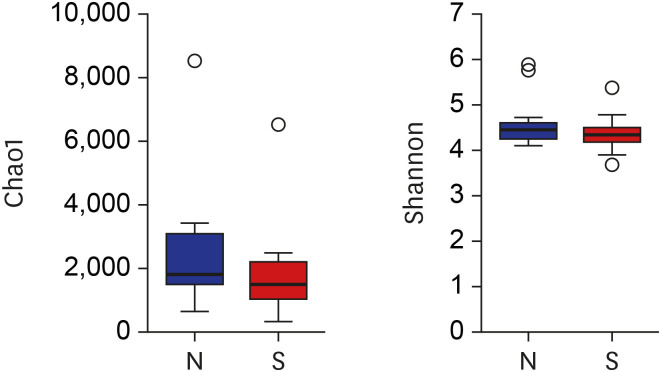
Raw reads were processed using the mothur software package (v1.39.4),
15 which carries out demultiplexing and denoising, quality filtering, and alignment. Operational taxonomic units (OTUs) were assigned to the reconstructed read sequences obtained from samples using the SILVA rRNA database (
http://www.mothur.org/wiki/Silva_reference_files), release 102 (
http://www.mothur.org/w/images/9/98/Silva.bacteria.zip). Chimeras were detected using Chimera UCHIME, and reads flagged as chimeras were submitted to the Ribosomal Database Project (RDP) reference database (release 16) classifier. The RDP reference database was used to assign taxonomic classifications to each OTU at the phylum, class, order, family, and genus levels. Other genetic materials, such as 18S rRNA gene fragments or 16S rRNA from Archaea, chloroplasts, and mitochondria present at this point in the process were removed. We resubmitted the effective sequences of each sample to the RDP Classifier to identify archaeal and bacterial sequences. Species richness and diversity statistics, including coverages, chao1, and Shannon indices, were also calculated using mothur. The Shapiro-Wilks test followed by parametric or nonparametric analysis was done before data analysis. We also used the Wilcoxon rank-sum test for two sample groups and the Kruskal-Wallis rank-sum test for multiple sample groups to identify significant differences in alpha diversity. Data visualization was performed using R (version 3.6.0),
16 and the graphics package
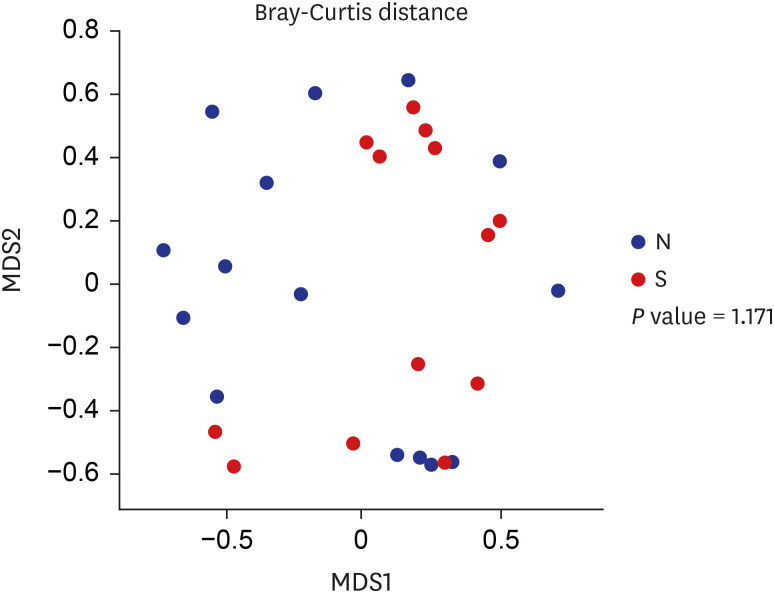
s MASS, ggplot2, and reshape2. To compare group beta diversities, we used non-metric multidimensional scaling (NMDS), which was performed using the phyloseq package for R. The linear discriminant analysis (LDA) effect size algorithm was used for putative bacterial identification with significant differential relative abundances. A default threshold value 2.0 on the logarithmic LDA score was used to identify discriminative features.
Ethics statement
The study protocol was approved by the Institutional Review Board of Inha University Hospital (INHAUH 2019-02-015) and requirement for informed consent was waived.
DISCUSSION
The existence of microbiota in healthy bile ducts is not widely accepted. The biliary system is typically maintained in a sterile condition by the continuous flushing action of bile and the bacteriostatic effects of bile salts. In addition, the sphincter of Oddi acts as an anatomical barrier that protects from bacterial invasion.
17 Therefore, under the conditions of normal bile flow, positive bile cultures are not expected, and the biliary microenvironment is believed to be hostile to most bacteria.
18 However, bacteria may remain, colonize, and replicate in the relatively stagnant bile environment created by biliary obstruction, and this would increase pressure within the biliary system. Several recent studies have reported relations between biliary microbiomes and biliary diseases such as cholangiocarcinoma and cholesterol gallstones, and high microbial diversity in the bile duct.
1319 Chen et al.
19 studies bile samples from 68 patients who required ERCP, and identified 252 bacterial genera and found the percentage compositions of the phyla Gemmatimonadetes, Nitrospirae, Chloro.exi, Latescibacteria, and Planctomycetes were higher in the bile of patients with distal cholangiocarcinoma than in those with other causes of biliary obstruction. They concluded that distal cholangiocarcinoma was associated with microbial compositional changes in the biliary tract.
19 Furthermore, in the study of Jiménez et al.,
20 the presence of intact bacteria in the bile duct was confirmed by microscopic examination within the biliary mucosa and up to 4.8 × 10
4 bacteria/ml of bile could be cultured from healthy pig. The evaluation of microbiota compositions in the bile duct in the general population is important in terms of elucidating the nature of relations between microbiota and specific diseases and determining the source of these biliary bacteria. However, due to ethical problems regarding the collection of bile from normal healthy individuals, it is difficult to determine the normal composition of biliary microbiota. Thus, we used bile juice from patients with various biliary diseases who required ERCP for the treatment of biliary obstruction, which is a limitation of the present study.
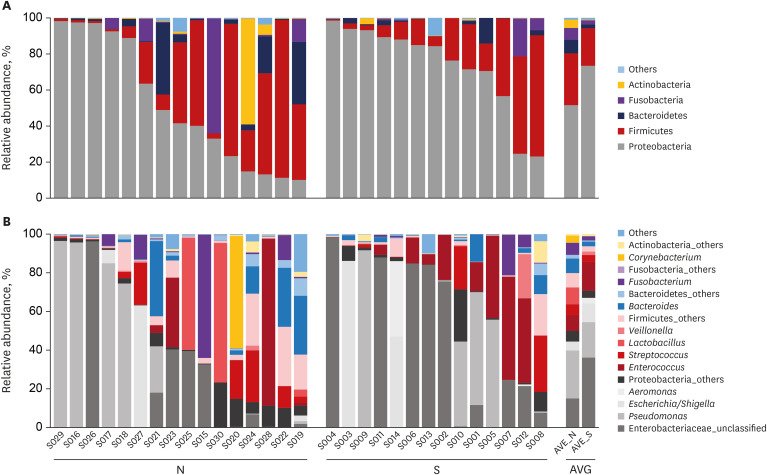
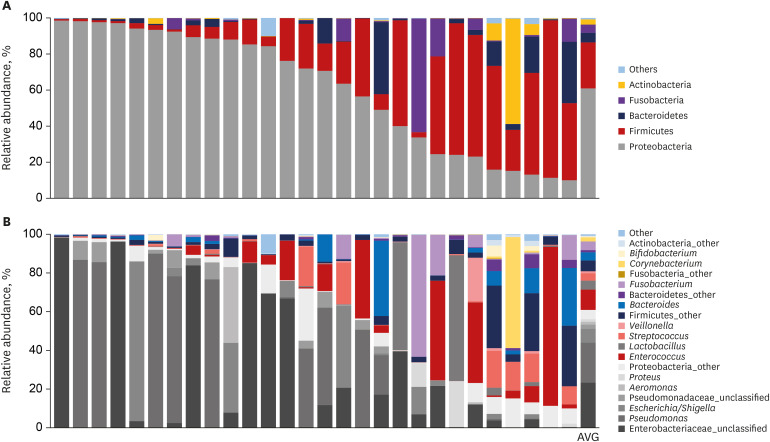
In the current study, an abundance of microbiota was found in the bile juice samples of patients with biliary obstruction. Although bile microbiota exhibited slight heterogeneity between individuals, Proteobacteria (61.7%) was the most dominant microbiota in almost all samples, and this value is similar to those reported in duodenum.
21 In the literature, Proteobacteria constitute ≤ 30% of phyla in the stomach, but ≤ 60% of phyla in the small intestine.
2122 Thus, our results provide strong support to the hypothesis that microbiota are present in the bile duct. Taken together, our results provide evidence that the duodenum is a primary source of biliary microbiota. The duodenum physiologically connects to the biliary system via the sphincter of Oddi; an anatomical barrier that prevents ascending infections from gut. The sphincter of Oddi regulates pressure in the biliary tract by controlling the excretions of pancreatic juice and bile, and thus, prevents duodenobiliary reflux. We suggest laxity of this mechanical barrier in patients with biliary diseases permits ascending reflux of intestinal bacteria, changes the biliary microenvironment, and facilitates the pathogeneses of biliary diseases. This suggestion is supported by a report issued by Ye et al.,
23 who investigated bacterial communities of the biliary tract, duodenum, stomach, and oral cavity in six patients with cholelithiasis by 16S rRNA amplicon sequencing. They reported that biliary microbiota compositions exhibited more similarity with duodenal microbiota than microbiota in other regions.
23 Since the aim of our study was to investigate biliary microbiota, we did not investigate retrograde infection of the biliary system by gut microbiota. Further studies are needed to clarify the mechanism involved.
Many authors have suggested that bacterial infection plays a role in the pathogenesis of choledocholithiasis, and culture and PCR studies have identified E. coli, K. pneumoniae, E. faecium, E. cloacae, and P. aeruginosa in bile, but no definitive causal relationship has been established between biliary bacteria and CBD stone formation.
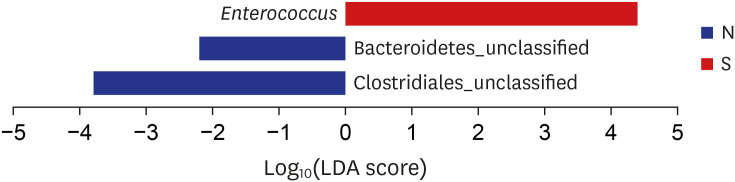
On comparing the compositions of biliary microbiomes in the CBD stone and control groups, we found the composition of
Enterococcus were found to be significantly different at the genus level (LDA score = 4.38;
P = 0.031), and thus, we suggest
Enterococcus may be associated with the development of pigment stones.
Enterococcus is a large genus of lactic acid bacteria of the phylum Firmicutes, and this genus is tolerant of a range of environmental conditions including a wide pH range (4.5–10.0), and high sodium chloride concentrations. Furthermore, microorganisms in this genus are common infecting organisms in acute cholangitis and malignant biliary obstruction.
24 Therefore, we assume that our results will be applicable to prevention of the recurrence rate of CBD stone after treatment with inducing elimination of
enterococcus in CBD by probiotics or antibiotics application. However, further clinical study dealing with this hypothesis would be required to use our results in the real clinical field.
Brown pigment stones are primarily composed of calcium salts of unconjugated bilirubinate, palmitate, carbonate, phosphate, and stearate.
625 The deconjugation of bile by bacterial beta-glucuronidase is the most commonly accepted mechanism for the formation of brown pigment stones. This enzyme is required for the deconjugation of bilirubin diglucuronide, which results in the release of free bilirubin and glucuronic acid,
6 and free bilirubin precipitates with Ca
2+ to form calcium bilirubinate, the major component of brown pigment stones.
26 According to a study by Leung et al.,
27
enterococcus is one of the beta-glucuronidase producing bacterial species isolated from biliary pigment stones and produces substantial amounts of beta-glucuronidase (the enzyme activity, 10.34 units). Therefore, this organism may be important in the deconjugation of bilirubin diglucuronide. Thus, we recommend that the mechanism whereby
enterococcus contributes to biliary stone formation be further investigated.
The present study has several limitations that warrant consideration. First, as mentioned above, our inability to enroll healthy normal controls is a study limitation. Second, the study is also limited by the small number of cases included, though sampling from patients using a nasobiliary tube is uncomfortable, and thus, we recruited only the minimum number of patients required for statistical analysis. Third, environmental conditions such as bile pH and the presence of suppurative cholangitis may have influenced microbiomes but were not measured. Fourth, we did not investigate the effects of specific microorganisms in bile on molecular or metabolomic changes in bile. Finally, there would be the risk of contamination of the upper gastrointestinal tract even if aspiration was achieved 24 hours after the endoscopic procedure. Therefore, the results in this study should be interpreted carefully.
In conclusion, we clearly demonstrated presence of bacteria in bile juice in bile duct obstruction patients, in which Proteobacteria predominate. Our results indicate that duodenal microbiota might influence the biliary microbiome in patients with biliary obstruction. Interestingly, we found that Enterococcus compositions in bile juice differed between patients with a brown pigment stone and controls, in which would suggest that the microbiome may be related with generation of pigment stones.
The main points of our study showed that the presence of bile microbiome in the patients with bile duct obstruction clearly, and demonstrated the composition of microbiome, in which Proteobacteria was predominant.
This composition of microbiome was quite similar with duodenal microbiome composition, and we suggest that duodenal microbiota might influence the biliary microbiome.
In the comparison results depending on the reason of bile duct obstruction, Enterococcus might be related to the development of brown pigment CBD stone.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download