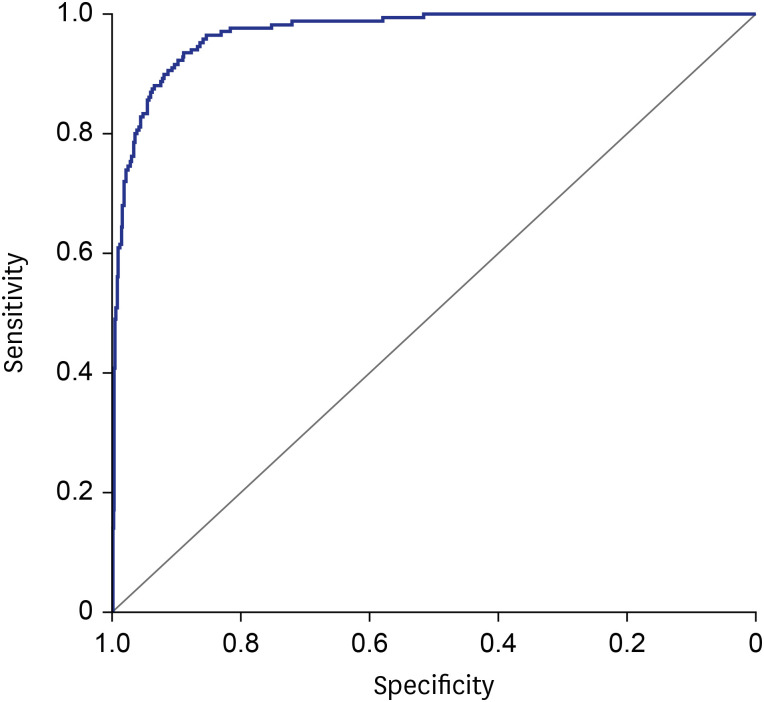
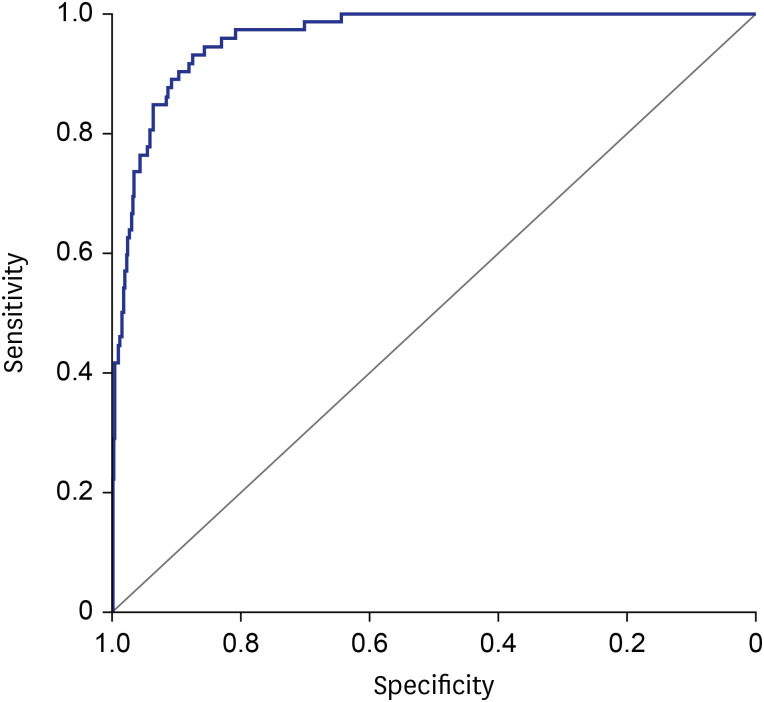
The 35 variables measured at hospital admission (
Table 2, see method) were included in the LASSO regression. After LASSO regression selection (
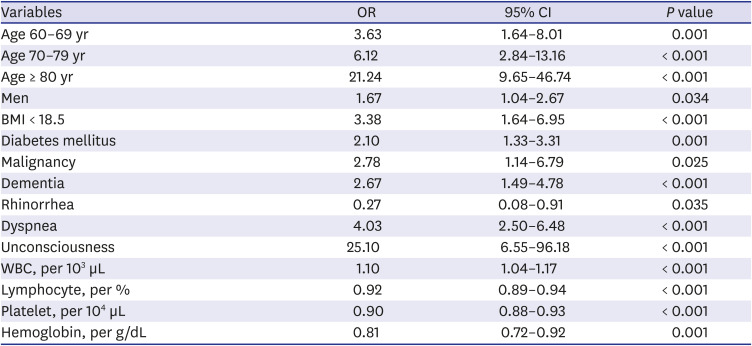
Supplementary Fig. 1), 17 variables remained as significant predictors of death, including age, sex, BMI, SBP, heart rate, body temperature, comorbidities including rhinorrhea, dyspnea, unconsciousness, diabetes mellitus, hypertension, chronic obstructive pulmonary disease, malignancy, WBC, lymphocyte, platelet, and hemoglobin. Inclusion of these 17 variables in a logistic regression model resulted in 13 variables that were independently statistically significant predictors of death and was included in risk score. These variables included age by three groups: 60–69 years (odds ratio [OR], 3.63; 95% CI, 1.64– 8.01;
P = 0.001), age 70–79 years (OR, 6.12; 95% CI, 2.84–13.16;
P < 0.001), age ≥ 80 years (OR, 21.24; 95% CI, 9.65–46.74;
P < 0.001), men (OR, 1.67; 95% CI, 1.04–2.67;
P = 0.034), BMI < 18.5 (OR, 3.38; 95% CI, 1.64–6.95;
P < 0.001), diabetes mellitus (OR, 2.10; 95% CI, 1.33–3.31;
P = 0.001), malignancy history (OR, 2.78; 95% CI, 1.14–6.79;
P = 0.025), dementia (OR, 2.67; 95% CI, 1.49–4.78;
P < 0.001), rhinorrhea (OR, 0.27; 95% CI, 0.08–0.91;
P = 0.035), dyspnea (OR, 4.03; 95% CI, 2.50–6.48;
P < 0.001), unconsciousness (OR, 25.10, 95% CI, 6.55–96.18;
P < 0.001), WBC (OR per 10
3 μL, 1.10; 95% CI, 1.04–1.17,
P < 0.001), lower lymphocyte proportion (OR per %, 0.92; 95% CI, 0.89–0.94;
P < 0.001), lower platelet count (OR per 10
4 μL, 0.90; 95% CI, 0.88–0.93,
P < 0.001), and lower hemoglobin level (OR per g/dL, 0.81; 95% CI, 0.72–0.92;
P = 0.001) (
Table 3).




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download