INTRODUCTION
METHODS
Animals and procedures
Cell culture and small interfering RNA (siRNA) transfection
Expression and purification of recombinant S100B protein
Histology
Immunohistochemistry
Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR)
Table 1
Primers for quantitative real-time reverse transcription polymerase chain reaction

Western blot analysis
Immunogold labeling and transmission electron microscopy analysis
Immunofluorescence staining
Statistical analyses
Ethics statement
RESULTS
Development of liver fibrosis in BDL mice
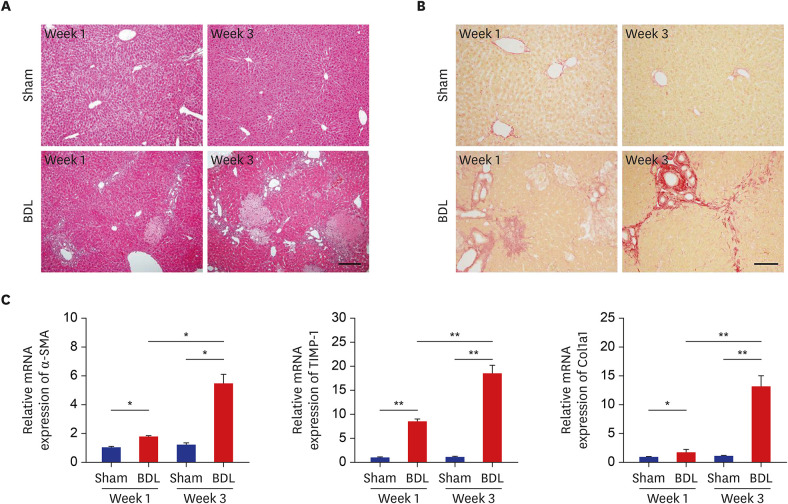 | Fig. 1Development of liver fibrosis after BDL. (A, B) Liver sections from sham control (upper panels) and BDL (lower panels) mice were examined by hematoxylin and eosin staining (A) and Sirius Red staining (B) at weeks 1 and 3 after BDL. (C) The mRNA levels of fibrotic marker genes at weeks 1 and 3 after BDL were determined by quantitative real-time reverse transcription polymerase chain reaction and normalized to β-actin (n = 9). The data represent the mean ± standard error of mean of three independent experiments. Scale bars, 25 μm.BDL = bile duct ligation, mRNA = messenger RNA, α-SMA = α-smooth muscle actin, TIMP-1 = tissue inhibitor of metalloproteinases-1, Col1a1 = collagen type I.
*P < 0.05, **P < 0.01.
|
Upregulation of S100B expression during liver fibrosis in BDL mice
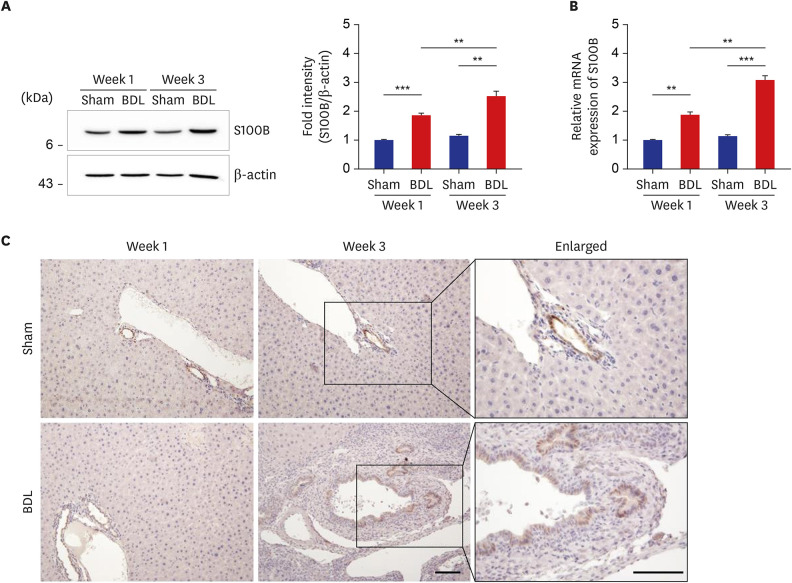 | Fig. 2Upregulated S100B expression in the livers of BDL mice. (A, B) Expression levels of S100B protein (A) and mRNA (B) in the livers from sham control and BDL mice was determined by Western blot analysis (A) and quantitative real-time reverse transcription polymerase chain reaction (B) and normalized to β-actin (n = 9) at each time point. The data represent the mean ± standard error of mean of three independent experiments. (C) Immunohistochemical staining of S100B in liver tissues from sham control and BDL mice. Note that an increased number of S100B-positive cells was found in the bile duct of BDL liver. Scale bars, 25 μm.BDL = bile duct ligation, mRNA = messenger RNA.
**P < 0.01, ***P < 0.001.
|
Increased expression of RAGE protein during liver fibrosis in BDL mice
 | Fig. 3Subcellular localization of S100B and RAGE in the livers of BDL mice. (A) Expression of RAGE protein in the livers of sham control and BDL mice at 1 week and 3 weeks after operation was analyzed by Western blot analysis (left panel) and normalized to β-actin (right panel) (n = 9). The data represent the mean ± standard error of mean of three independent experiments. (B, C) Triple immunofluorescence staining of S100B, RAGE, and α-SMA (B) or CK19 (C) were performed in the liver sections of sham control and BDL mice at 3 weeks after operation. Note the colocalization of S100B and RAGE in α-SMA-positive activated HSCs (B) as well as CK19-positive bile duct epithelial cells (C) in the BDL livers. Green, S100B; red, α-SMA, and CK19; white, RAGE. Scale bars, 20 μm.RAGE = receptor for advanced glycation end products, BDL = bile duct ligation, α-SMA = α-smooth muscle actin, CK19 = cytokeratin19.
*P < 0.05, **P < 0.01, ***P < 0.001.
|
Colocalization of S100B and RAGE in activated HSCs as well as bile duct epithelial cells in the livers of BDL mice
 | Fig. 4Double immunogold labeling of S100B and RAGE in the livers of BDL mice. The liver sections from sham control and BDL mice at week 3 were incubated with rabbit monoclonal anti-S100B (1:100) and mouse monoclonal anti-RAGE (1:100), followed by incubation with goat anti-rabbit IgG-conjugated 15 nm gold particles for S100B and goat anti-mouse IgG-conjugated 25 nm gold particles for RAGE. Note that immunogold particles indicating S100B (arrows) and RAGE (arrowhead) are present in the cytoplasm and microvilli of bile duct epithelial cells (upper panels), and in the HSCs, both particles were found in the nucleus and its environs (lower panels). Scale bars, 2 μm.RAGE = receptor for advanced glycation end products, BDL = bile duct ligation, HSC = hepatic stellate cell, IgG = immunoglobulin G, N = nuclei, M = mitochondria, L = lumen, LD = lipid droplet.
|
S100B regulates HSC activation through RAGE
 | Fig. 5S100B regulates HSC activation via RAGE. (A, B) HSC-T6 cells were treated with recombinant mouse S100B protein in a dose-dependent manner. After 12 hours, the cells were lysed, and HSC activation-related protein and gene expression levels were detected by Western blotting (A) and qRT-PCR (B), respectively. (C, D) HSC-T6 cells were transiently transfected with either SCR or si-RAGE in a dose-dependent manner, then incubated with 25 ng of recombinant S100B protein for 12 hours and analyzed by Western blotting (C) and qRT-PCR (D). The data represent the mean ± standard error of mean of three independent experiments.RAGE = receptor for advanced glycation end products, Col1a1 = collagen type I, α-SMA = α-smooth muscle actin, GAPDH = glyceraldehyde 3-phosphate dehydrogenase, mRNA = messenger RNA, PBS = phosphate-buffered saline, SCR = scrambled, si-RAGE = small interfering RNA-mediated knockdown of endogenous RAGE, qRT-PCR = quantitative real-time reverse transcription polymerase chain reaction, HSC = hepatic stellate cell.
*P < 0.05, **P < 0.01, ***P < 0.001.
|




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download